Fine-mapping inflammatory bowel disease loci to single-variant resolution
- PMID: 28658209
- PMCID: PMC5511510
- DOI: 10.1038/nature22969
Fine-mapping inflammatory bowel disease loci to single-variant resolution
Abstract
Inflammatory bowel diseases are chronic gastrointestinal inflammatory disorders that affect millions of people worldwide. Genome-wide association studies have identified 200 inflammatory bowel disease-associated loci, but few have been conclusively resolved to specific functional variants. Here we report fine-mapping of 94 inflammatory bowel disease loci using high-density genotyping in 67,852 individuals. We pinpoint 18 associations to a single causal variant with greater than 95% certainty, and an additional 27 associations to a single variant with greater than 50% certainty. These 45 variants are significantly enriched for protein-coding changes (n = 13), direct disruption of transcription-factor binding sites (n = 3), and tissue-specific epigenetic marks (n = 10), with the last category showing enrichment in specific immune cells among associations stronger in Crohn's disease and in gut mucosa among associations stronger in ulcerative colitis. The results of this study suggest that high-resolution fine-mapping in large samples can convert many discoveries from genome-wide association studies into statistically convincing causal variants, providing a powerful substrate for experimental elucidation of disease mechanisms.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
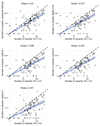





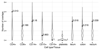
References
-
- Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42. quiz e30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA141743/CA/NCI NIH HHS/United States
- U54 DE023789/DE/NIDCR NIH HHS/United States
- U01 DK062413/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- U24 DK062429/DK/NIDDK NIH HHS/United States
- U01 DK062432/DK/NIDDK NIH HHS/United States
- MR/M00533X/1/MRC_/Medical Research Council/United Kingdom
- U01 DK062429/DK/NIDDK NIH HHS/United States
- 098051/WT_/Wellcome Trust/United Kingdom
- U01 DK062422/DK/NIDDK NIH HHS/United States
- G0800759/MRC_/Medical Research Council/United Kingdom
- ETM/137/CSO_/Chief Scientist Office/United Kingdom
- R01 DK106593/DK/NIDDK NIH HHS/United States
- U01 DK062420/DK/NIDDK NIH HHS/United States
- R01 DK064869/DK/NIDDK NIH HHS/United States
- 098759/WT_/Wellcome Trust/United Kingdom
- U01 AI067068/AI/NIAID NIH HHS/United States
- P01 DK046763/DK/NIDDK NIH HHS/United States
- R01 DK092235/DK/NIDDK NIH HHS/United States
- G0600329/MRC_/Medical Research Council/United Kingdom

