In Vivo Regulation of the Zebrafish Endoderm Progenitor Niche by T-Box Transcription Factors
- PMID: 28658625
- PMCID: PMC5494305
- DOI: 10.1016/j.celrep.2017.06.011
In Vivo Regulation of the Zebrafish Endoderm Progenitor Niche by T-Box Transcription Factors
Abstract
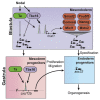
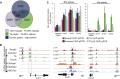
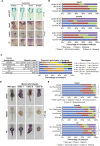
T-box transcription factors T/Brachyury homolog A (Ta) and Tbx16 are essential for correct mesoderm development in zebrafish. The downstream transcriptional networks guiding their functional activities are poorly understood. Additionally, important contributions elsewhere are likely masked due to redundancy. Here, we exploit functional genomic strategies to identify Ta and Tbx16 targets in early embryogenesis. Surprisingly, we discovered they not only activate mesodermal gene expression but also redundantly regulate key endodermal determinants, leading to substantial loss of endoderm in double mutants. To further explore the gene regulatory networks (GRNs) governing endoderm formation, we identified targets of Ta/Tbx16-regulated homeodomain transcription factor Mixl1, which is absolutely required in zebrafish for endoderm formation. Interestingly, we find many endodermal determinants coordinately regulated through common genomic occupancy by Mixl1, Eomesa, Smad2, Nanog, Mxtx2, and Pou5f3. Collectively, these findings augment the endoderm GRN and reveal a panel of target genes underlying the Ta, Tbx16, and Mixl1 mutant phenotypes.
Keywords: ChIP-seq; T-box; endoderm; redundancy; transcription.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Global identification of Smad2 and Eomesodermin targets in zebrafish identifies a conserved transcriptional network in mesendoderm and a novel role for Eomesodermin in repression of ectodermal gene expression.BMC Biol. 2014 Oct 3;12:81. doi: 10.1186/s12915-014-0081-5. BMC Biol. 2014. PMID: 25277163 Free PMC article.
-
Maternal Eomesodermin regulates zygotic nodal gene expression for mesendoderm induction in zebrafish embryos.J Mol Cell Biol. 2014 Aug;6(4):272-85. doi: 10.1093/jmcb/mju028. Epub 2014 Jun 12. J Mol Cell Biol. 2014. PMID: 24924767
-
tbx6l and tbx16 are redundantly required for posterior paraxial mesoderm formation during zebrafish embryogenesis.Dev Dyn. 2017 Oct;246(10):759-769. doi: 10.1002/dvdy.24547. Epub 2017 Aug 30. Dev Dyn. 2017. PMID: 28691257 Free PMC article.
-
Molecular basis of vertebrate endoderm development.Int Rev Cytol. 2007;259:49-111. doi: 10.1016/S0074-7696(06)59002-3. Int Rev Cytol. 2007. PMID: 17425939 Review.
-
Molecular regulation of vertebrate early endoderm development.Dev Biol. 2002 Sep 15;249(2):191-203. doi: 10.1006/dbio.2002.0765. Dev Biol. 2002. PMID: 12221001 Review.
Cited by
-
Long-Range Signaling Activation and Local Inhibition Separate the Mesoderm and Endoderm Lineages.Dev Cell. 2018 Jan 22;44(2):179-191.e5. doi: 10.1016/j.devcel.2017.11.021. Epub 2017 Dec 21. Dev Cell. 2018. PMID: 29275993 Free PMC article.
-
FoxH1 represses miR-430 during early embryonic development of zebrafish via non-canonical regulation.BMC Biol. 2019 Jul 30;17(1):61. doi: 10.1186/s12915-019-0683-z. BMC Biol. 2019. PMID: 31362746 Free PMC article.
-
A conserved regulatory program initiates lateral plate mesoderm emergence across chordates.Nat Commun. 2019 Aug 26;10(1):3857. doi: 10.1038/s41467-019-11561-7. Nat Commun. 2019. PMID: 31451684 Free PMC article.
-
Nodal signaling establishes a competency window for stochastic cell fate switching.Dev Cell. 2022 Dec 5;57(23):2604-2622.e5. doi: 10.1016/j.devcel.2022.11.008. Dev Cell. 2022. PMID: 36473458 Free PMC article.
-
Pluripotency factors determine gene expression repertoire at zygotic genome activation.Nat Commun. 2022 Feb 10;13(1):788. doi: 10.1038/s41467-022-28434-1. Nat Commun. 2022. PMID: 35145080 Free PMC article.
References
-
- Ahn D., You K.H., Kim C.H. Evolution of the tbx6/16 subfamily genes in vertebrates: insights from zebrafish. Mol. Biol. Evol. 2012;29:3959–3983. - PubMed
-
- Amacher S.L., Draper B.W., Summers B.R., Kimmel C.B. The zebrafish T-box genes no tail and spadetail are required for development of trunk and tail mesoderm and medial floor plate. Development. 2002;129:3311–3323. - PubMed
-
- Amack J.D., Wang X., Yost H.J. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev. Biol. 2007;310:196–210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

