The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles
- PMID: 28659343
- PMCID: PMC5566525
- DOI: 10.1074/jbc.M117.797332
The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles
Erratum in
-
Correction: The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles.J Biol Chem. 2019 Dec 6;294(49):18951. doi: 10.1074/jbc.AAC119.011798. J Biol Chem. 2019. PMID: 31811051 Free PMC article. No abstract available.
Abstract
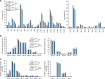
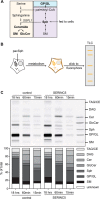
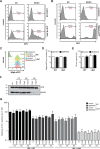
The host-cell restriction factor SERINC5 potently suppresses the infectivity of HIV, type 1 (HIV-1) particles, and is counteracted by the viral pathogenesis factor Nef. However, the molecular mechanism by which SERINC5 restricts HIV-1 particle infectivity is still unclear. Because SERINC proteins have been suggested to facilitate the incorporation of serine during the biosynthesis of membrane lipids and because lipid composition of HIV particles is a major determinant of the infectious potential of the particles, we tested whether SERINC5-mediated restriction of HIV particle infectivity involves alterations of membrane lipid composition. We produced and purified HIV-1 particles from SERINC5293T cells with very low endogenous SERINC5 levels under conditions in which ectopically expressed SERINC5 restricts HIV-1 infectivity and is antagonized by Nef and analyzed both virions and producer cells with quantitative lipid MS. SERINC5 restriction and Nef antagonism were not associated with significant alterations in steady-state lipid composition of producer cells and HIV particles. Sphingosine metabolism kinetics were also unaltered by SERINC5 expression. Moreover, the levels of phosphatidylserine on the surface of HIV-1 particles, which may trigger uptake into non-productive internalization pathways in target cells, did not change upon expression of SERINC5 or Nef. Finally, saturating the phosphatidylserine-binding sites on HIV target cells did not affect SERINC5 restriction or Nef antagonism. These results demonstrate that the restriction of HIV-1 particle infectivity by SERINC5 does not depend on alterations in lipid composition and organization of HIV-1 particles and suggest that channeling serine into lipid biosynthesis may not be a cardinal cellular function of SERINC5.
Keywords: Nef; SERINC5; host cell restriction factor; host-pathogen interaction; human immunodeficiency virus (HIV); infectious disease; lipid; lipid composition; viral protein.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Altfeld M., and Gale M. Jr. (2015) Innate immunity against HIV-1 infection. Nat. Immunol. 16, 554–562 - PubMed
-
- Laguette N., Brégnard C., Hue P., Basbous J., Yatim A., Larroque M., Kirchhoff F., Constantinou A., Sobhian B., and Benkirane M. (2014) Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156, 134–145 - PubMed
-
- Neil S. J., Zang T., and Bieniasz P. D. (2008) Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

