CEP19 cooperates with FOP and CEP350 to drive early steps in the ciliogenesis programme
- PMID: 28659385
- PMCID: PMC5493781
- DOI: 10.1098/rsob.170114
CEP19 cooperates with FOP and CEP350 to drive early steps in the ciliogenesis programme
Abstract
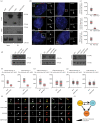
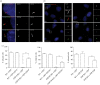
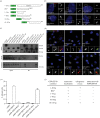
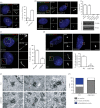
Primary cilia are microtubule-based sensory organelles necessary for efficient transduction of extracellular cues. To initiate cilia formation, ciliary vesicles (CVs) are transported to the vicinity of the centrosome where they dock to the distal end of the mother centriole and fuse to initiate cilium assembly. However, to this date, the early steps in cilia formation remain incompletely understood. Here, we demonstrate functional interplay between CEP19, FOP and CEP350 in ciliogenesis. Using three-dimensional structured-illumination microscopy (3D-SIM) imaging, we mapped the relative spatial distribution of these proteins at the distal end of the mother centriole and show that CEP350/FOP act upstream of CEP19 in their recruitment hierarchy. We demonstrate that CEP19 CRISPR KO cells are severely impaired in their ability to form cilia, analogous to the loss of function of CEP19 binding partners FOP and CEP350. Notably, in the absence of CEP19 microtubule anchoring at centromes is similar in manner to its interaction partners FOP and CEP350. Using GFP-tagged deletion constructs of CEP19, we show that the C-terminus of CEP19 is required for both its localization to centrioles and for its function in ciliogenesis. Critically, this region also mediates the interaction between CEP19 and FOP/CEP350. Interestingly, a morbid-obesity-associated R82* truncated mutant of CEP19 cannot ciliate nor interact with FOP and CEP350, indicative of a putative role for CEP19 in ciliopathies. Finally, analysis of CEP19 KO cells using thin-section electron microscopy revealed marked defects in the docking of CVs to the distal end of the mother centrioles. Together, these data demonstrate a role for the CEP19, FOP and CEP350 module in ciliogenesis and the possible effect of disrupting their functions in ciliopathies.
Keywords: centrioles; centrosomes; cilia; ciliopathies; super-resolution microscopy.
© 2017 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures

References
-
- Bornens M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25–34. (doi:10.1016/S0955-0674(01)00290-3) - DOI - PubMed
-
- Anderson RGW. 1972. The three-dimensional structure of the basal body from the rhesus monkey oviiduct. J. Cell Biol. 54, 246–265. (doi:10.1083/jcb.54.2.246) - DOI - PMC - PubMed
-
- Eggenschwiler JT, Anderson KV. 2007. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345–373. (doi:10.1146/annurev.cellbio.23.090506.123249) - DOI - PMC - PubMed
-
- Vorobjev IA, Chentsov Yu S. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93, 938 (doi:10.1083/jcb.93.3.938) - DOI - PMC - PubMed
-
- Reiter JF, Blacque OE, Leroux MR. 2012. The base of the cilium: roles for transition fibers and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13, 608–618. (doi:10.1038/embor.2012.73) - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

