Enfuvirtide (T20)-Based Lipopeptide Is a Potent HIV-1 Cell Fusion Inhibitor: Implications for Viral Entry and Inhibition
- PMID: 28659478
- PMCID: PMC5571253
- DOI: 10.1128/JVI.00831-17
Enfuvirtide (T20)-Based Lipopeptide Is a Potent HIV-1 Cell Fusion Inhibitor: Implications for Viral Entry and Inhibition
Abstract
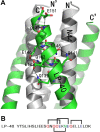
The peptide drug enfuvirtide (T20) is the only viral fusion inhibitor used in combination therapy for HIV-1 infection, but it has relatively low antiviral activity and easily induces drug resistance. Emerging studies demonstrate that lipopeptide-based fusion inhibitors, such as LP-11 and LP-19, which mainly target the gp41 pocket site, have greatly improved antiviral potency and in vivo stability. In this study, we focused on developing a T20-based lipopeptide inhibitor that lacks pocket-binding sequence and targets a different site. First, the C-terminal tryptophan-rich motif (TRM) of T20 was verified to be essential for its target binding and inhibition; then, a novel lipopeptide, termed LP-40, was created by replacing the TRM with a fatty acid group. LP-40 showed markedly enhanced binding affinity for the target site and dramatically increased inhibitory activity on HIV-1 membrane fusion, entry, and infection. Unlike LP-11 and LP-19, which required a flexible linker between the peptide sequence and the lipid moiety, addition of a linker to LP-40 sharply reduced its potency, implying different binding modes with the extended N-terminal helices of gp41. Also, interestingly, LP-40 showed more potent activity than LP-11 in inhibiting HIV-1 Env-mediated cell-cell fusion while it was less active than LP-11 in inhibiting pseudovirus entry, and the two inhibitors displayed synergistic antiviral effects. The crystal structure of LP-40 in complex with a target peptide revealed their key binding residues and motifs. Combined, our studies have not only provided a potent HIV-1 fusion inhibitor, but also revealed new insights into the mechanisms of viral inhibition.IMPORTANCE T20 is the only membrane fusion inhibitor available for treatment of viral infection; however, T20 requires high doses and has a low genetic barrier for resistance, and its inhibitory mechanism and structural basis remain unclear. Here, we report the design of LP-40, a T20-based lipopeptide inhibitor that has greatly improved anti-HIV activity and is a more potent inhibitor of cell-cell fusion than of cell-free virus infection. The binding modes of two classes of membrane-anchoring lipopeptides (LP-40 and LP-11) verify the current fusion model in which an extended prehairpin structure bridges the viral and cellular membranes, and their complementary effects suggest a vital strategy for combination therapy of HIV-1 infection. Moreover, our understanding of the mechanism of action of T20 and its derivatives benefits from the crystal structure of LP-40.
Keywords: HIV-1; T20; fusion inhibitor; gp41; lipopeptide.
Copyright © 2017 American Society for Microbiology.
Figures








Similar articles
-
A Lipopeptide HIV-1/2 Fusion Inhibitor with Highly Potent In Vitro, Ex Vivo, and In Vivo Antiviral Activity.J Virol. 2017 May 12;91(11):e00288-17. doi: 10.1128/JVI.00288-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28356533 Free PMC article.
-
Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus.J Biol Chem. 2018 Apr 6;293(14):5323-5334. doi: 10.1074/jbc.RA118.001729. Epub 2018 Feb 7. J Biol Chem. 2018. PMID: 29425101 Free PMC article.
-
Design and Characterization of Cholesterylated Peptide HIV-1/2 Fusion Inhibitors with Extremely Potent and Long-Lasting Antiviral Activity.J Virol. 2019 May 15;93(11):e02312-18. doi: 10.1128/JVI.02312-18. Print 2019 Jun 1. J Virol. 2019. PMID: 30867304 Free PMC article.
-
Biochemistry and biophysics of HIV-1 gp41 - membrane interactions and implications for HIV-1 envelope protein mediated viral-cell fusion and fusion inhibitor design.Curr Top Med Chem. 2011 Dec;11(24):2959-84. doi: 10.2174/156802611798808497. Curr Top Med Chem. 2011. PMID: 22044229 Free PMC article. Review.
-
Inhibition of HIV-1 by fusion inhibitors.Curr Pharm Des. 2010;16(33):3716-28. doi: 10.2174/138161210794079218. Curr Pharm Des. 2010. PMID: 21128887 Review.
Cited by
-
TLR10 Senses HIV-1 Proteins and Significantly Enhances HIV-1 Infection.Front Immunol. 2019 Mar 15;10:482. doi: 10.3389/fimmu.2019.00482. eCollection 2019. Front Immunol. 2019. PMID: 30930906 Free PMC article.
-
Conserved Residue Asn-145 in the C-Terminal Heptad Repeat Region of HIV-1 gp41 is Critical for Viral Fusion and Regulates the Antiviral Activity of Fusion Inhibitors.Viruses. 2019 Jul 3;11(7):609. doi: 10.3390/v11070609. Viruses. 2019. PMID: 31277353 Free PMC article.
-
Helical sulfonyl-γ-AApeptides for the inhibition of HIV-1 fusion and HIF-1α signaling.RSC Med Chem. 2024 Mar 20;15(5):1418-1423. doi: 10.1039/d4md00110a. eCollection 2024 May 22. RSC Med Chem. 2024. PMID: 38784464 Free PMC article. Review.
-
Monotherapy with a low-dose lipopeptide HIV fusion inhibitor maintains long-term viral suppression in rhesus macaques.PLoS Pathog. 2019 Feb 4;15(2):e1007552. doi: 10.1371/journal.ppat.1007552. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30716118 Free PMC article.
-
Will Peptides Help to Stop COVID-19?Biochemistry (Mosc). 2022 Jul;87(7):590-604. doi: 10.1134/S0006297922070021. Biochemistry (Mosc). 2022. PMID: 36154880 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

