Replications of Two Closely Related Groups of Jumbo Phages Show Different Level of Dependence on Host-encoded RNA Polymerase
- PMID: 28659872
- PMCID: PMC5468394
- DOI: 10.3389/fmicb.2017.01010
Replications of Two Closely Related Groups of Jumbo Phages Show Different Level of Dependence on Host-encoded RNA Polymerase
Abstract
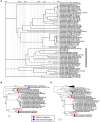
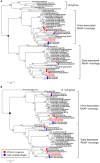
Ralstonia solanacearum phages ΦRP12 and ΦRP31 are jumbo phages isolated in Thailand. Here we show that they exhibit similar virion morphology, genome organization and host range. Genome comparisons as well as phylogenetic and proteomic tree analyses support that they belong to the group of ΦKZ-related phages, with their closest relatives being R. solanacearum phages ΦRSL2 and ΦRSF1. Compared with ΦRSL2 and ΦRSF1, ΦRP12 and ΦRP31 possess larger genomes (ca. 280 kbp, 25% larger). The replication of ΦRP12 and ΦRP31 was not affected by rifampicin treatment (20 μg/ml), suggesting that phage-encoded RNAPs function to start and complete the infection cycle of these phages without the need of host-encoded RNAPs. In contrast, ΦRSL2 and ΦRSF1, encoding the same set of RNAPs, did not produce progeny phages in the presence of rifampicin (5 μg/ml). This observation opens the possibility that some ΦRP12/ΦRP31 factors that are absent in ΦRSL2 and ΦRSF1 are involved in their host-independent transcription.
Keywords: Ralstonia solanacearum; genomic analysis; jumbo phages; virion-associated-RNA polymerase; ΦKZ-like phages.
Figures


References
-
- Bhunchoth A., Phironrit N., Leksomboon C., Chatchawankanphanich O., Kotera S., Narulita E., et al. (2015). Isolation of Ralstonia solanacearum-infecting bacteriophages from tomato fields in Chiang Mai, Thailand, and their experimetal use as biocontrol agents. J. Appl. Microbiol. 118, 1023–1033. 10.1111/jam.12763 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

