Comparison of Biochemical Properties of HIV-1 and HIV-2 Capsid Proteins
- PMID: 28659897
- PMCID: PMC5469281
- DOI: 10.3389/fmicb.2017.01082
Comparison of Biochemical Properties of HIV-1 and HIV-2 Capsid Proteins
Abstract
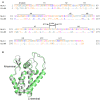
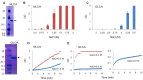
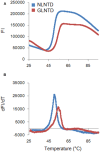
Timely disassembly of viral core composed of self-assembled capsid (CA) in infected host cells is crucial for retroviral replication. Extensive in vitro studies to date on the self-assembly/disassembly mechanism of human immunodeficiency virus type 1 (HIV-1) CA have revealed its core structure and amino acid residues essential for CA-CA intermolecular interaction. However, little is known about in vitro properties of HIV-2 CA. In this study, we comparatively analyzed the polymerization properties of bacterially expressed HIV-1 and HIV-2 CA proteins. Interestingly, a much higher concentration of NaCl was required for HIV-2 CA to self-assemble than that for HIV-1 CA, but once the polymerization started, the reaction proceeded more rapidly than that observed for HIV-1 CA. Analysis of a chimeric protein revealed that N-terminal domain (NTD) is responsible for this unique property of HIV-2 CA. To further study the molecular basis for different in vitro properties of HIV-1 and HIV-2 CA proteins, we determined thermal stabilities of HIV-1 and HIV-2 CA NTD proteins at several NaCl concentrations by fluorescent-based thermal shift assays. Experimental data obtained showed that HIV-2 CA NTD was structurally more stable than HIV-1 CA NTD. Taken together, our results imply that distinct in vitro polymerization abilities of the two CA proteins are related to their structural instability/stability, which is one of the decisive factors for viral replication potential. In addition, our assay system described here may be potentially useful for searching for anti-CA antivirals against HIV-1 and HIV-2.
Keywords: CA-polymerization; CA-stability; Gag-CA; HIV-1; HIV-2; NTD.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

