Transcription Factor Repertoire of Necrotrophic Fungal Phytopathogen Ascochyta rabiei: Predominance of MYB Transcription Factors As Potential Regulators of Secretome
- PMID: 28659964
- PMCID: PMC5470089
- DOI: 10.3389/fpls.2017.01037
Transcription Factor Repertoire of Necrotrophic Fungal Phytopathogen Ascochyta rabiei: Predominance of MYB Transcription Factors As Potential Regulators of Secretome
Abstract
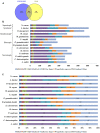
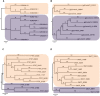
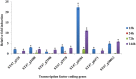
Transcription factors (TFs) are the key players in gene expression and their study is highly significant for shedding light on the molecular mechanisms and evolutionary history of organisms. During host-pathogen interaction, extensive reprogramming of gene expression facilitated by TFs is likely to occur in both host and pathogen. To date, the knowledge about TF repertoire in filamentous fungi is in infancy. The necrotrophic fungus Ascochyta rabiei, that causes destructive Ascochyta blight (AB) disease of chickpea (Cicer arietinum), demands more comprehensive study for better understanding of Ascochyta-legume pathosystem. In the present study, we performed the genome-wide identification and analysis of TFs in A. rabiei. Taking advantage of A. rabiei genome sequence, we used a bioinformatic approach to predict the TF repertoire of A. rabiei. For identification and classification of A. rabiei TFs, we designed a comprehensive pipeline using a combination of BLAST and InterProScan software. A total of 381 A. rabiei TFs were predicted and divided into 32 fungal specific families of TFs. The gene structure, domain organization and phylogenetic analysis of abundant families of A. rabiei TFs were also carried out. Comparative study of A. rabiei TFs with that of other necrotrophic, biotrophic, hemibiotrophic, symbiotic, and saprotrophic fungi was performed. It suggested presence of both conserved as well as unique features among them. Moreover, cis-acting elements on promoter sequences of earlier predicted A. rabiei secretome were also identified. With the help of published A. rabiei transcriptome data, the differential expression of TF and secretory protein coding genes was analyzed. Furthermore, comprehensive expression analysis of few selected A. rabiei TFs using quantitative real-time polymerase chain reaction revealed variety of expression patterns during host colonization. These genes were expressed in at least one of the time points tested post infection. Overall, this study illustrates the first genome-wide identification and analysis of TF repertoire of A. rabiei. This work would provide the basis for further studies to dissect role of TFs in the molecular mechanisms during A. rabiei-chickpea interactions.
Keywords: Ascochyta rabiei; cis-acting elements; necrotrophic fungi; plant–pathogen interaction; secretome; transcription factors.
Figures








References
-
- Adryan B., Teichmann S. A. (2006). FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics 22 1532–1533. - PubMed
-
- Asano Y., Hagiwara D., Yamashino T., Mizuno T. (2007). Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71 1800–1803. - PubMed
-
- Bok J. W., Wiemann P., Garvey G. S., Lim F. Y., Haas B., Wortman J., et al. (2014). Illumina identification of RsrA, a conserved C2H2 transcription factor coordinating the NapA mediated oxidative stress signaling pathway in Aspergillus. BMC Genomics 15:1011 10.1186/1471-2164-15-1011 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

