A Mighty "Protein Extractor" of the Cell: Structure and Function of the p97/CDC48 ATPase
- PMID: 28660197
- PMCID: PMC5468458
- DOI: 10.3389/fmolb.2017.00039
A Mighty "Protein Extractor" of the Cell: Structure and Function of the p97/CDC48 ATPase
Abstract
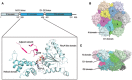
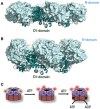
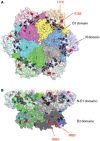
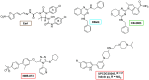
p97/VCP (known as Cdc48 in S. cerevisiae or TER94 in Drosophila) is one of the most abundant cytosolic ATPases. It is highly conserved from archaebacteria to eukaryotes. In conjunction with a large number of cofactors and adaptors, it couples ATP hydrolysis to segregation of polypeptides from immobile cellular structures such as protein assemblies, membranes, ribosome, and chromatin. This often results in proteasomal degradation of extracted polypeptides. Given the diversity of p97 substrates, this "segregase" activity has profound influence on cellular physiology ranging from protein homeostasis to DNA lesion sensing, and mutations in p97 have been linked to several human diseases. Here we summarize our current understanding of the structure and function of this important cellular machinery and discuss the relevant clinical implications.
Keywords: AAA ATPase; Cdc48; chaperones; neurodegenerative diseases; p97/VCP; protein denaturation; protein quality control.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

