METTL21B Is a Novel Human Lysine Methyltransferase of Translation Elongation Factor 1A: Discovery by CRISPR/Cas9 Knockout
- PMID: 28663172
- PMCID: PMC5724183
- DOI: 10.1074/mcp.M116.066308
METTL21B Is a Novel Human Lysine Methyltransferase of Translation Elongation Factor 1A: Discovery by CRISPR/Cas9 Knockout
Abstract
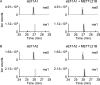
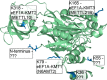
Lysine methylation is widespread on human proteins, however the enzymes that catalyze its addition remain largely unknown. This limits our capacity to study the function and regulation of this modification. Here we used the CRISPR/Cas9 system to knockout putative protein methyltransferases METTL21B and METTL23 in K562 cells, to determine if they methylate elongation factor eEF1A. The known eEF1A methyltransferase EEF1AKMT1 was also knocked out as a control. Targeted mass spectrometry revealed the loss of lysine 165 methylation upon knockout of METTL21B, and the expected loss of lysine 79 methylation on knockout of EEF1AKMT1 No loss of eEF1A methylation was seen in the METTL23 knockout. Recombinant METTL21B was shown in vitro to catalyze methylation on lysine 165 in eEF1A1 and eEF1A2, confirming it as the methyltransferase responsible for this methylation site. Proteomic analysis by SILAC revealed specific upregulation of large ribosomal subunit proteins in the METTL21B knockout, and changes to further processes related to eEF1A function in knockouts of both METTL21B and EEF1AKMT1 This indicates that the methylation of lysine 165 in human eEF1A has a very specific role. METTL21B exists only in vertebrates, with its target lysine showing similar evolutionary conservation. We suggest METTL21B be renamed eEF1A-KMT3. This is the first study to specifically generate CRISPR/Cas9 knockouts of putative protein methyltransferase genes, for substrate discovery and site mapping. Our approach should prove useful for the discovery of further novel methyltransferases, and more generally for the discovery of sites for other protein-modifying enzymes.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A).Nucleic Acids Res. 2017 May 5;45(8):4370-4389. doi: 10.1093/nar/gkx002. Nucleic Acids Res. 2017. PMID: 28108655 Free PMC article.
-
Novel N-terminal and Lysine Methyltransferases That Target Translation Elongation Factor 1A in Yeast and Human.Mol Cell Proteomics. 2016 Jan;15(1):164-76. doi: 10.1074/mcp.M115.052449. Epub 2015 Nov 6. Mol Cell Proteomics. 2016. PMID: 26545399 Free PMC article.
-
Saccharomyces cerevisiae Eukaryotic Elongation Factor 1A (eEF1A) Is Methylated at Lys-390 by a METTL21-Like Methyltransferase.PLoS One. 2015 Jun 26;10(6):e0131426. doi: 10.1371/journal.pone.0131426. eCollection 2015. PLoS One. 2015. PMID: 26115316 Free PMC article.
-
Methylation of Elongation Factor 1A: Where, Who, and Why?Trends Biochem Sci. 2018 Mar;43(3):211-223. doi: 10.1016/j.tibs.2018.01.004. Epub 2018 Feb 1. Trends Biochem Sci. 2018. PMID: 29398204 Review.
-
Structure and function of DNA methyltransferases.Annu Rev Biophys Biomol Struct. 1995;24:293-318. doi: 10.1146/annurev.bb.24.060195.001453. Annu Rev Biophys Biomol Struct. 1995. PMID: 7663118 Review.
Cited by
-
Methyltransferase-like 21e inhibits 26S proteasome activity to facilitate hypertrophy of type IIb myofibers.FASEB J. 2019 Aug;33(8):9672-9684. doi: 10.1096/fj.201900582R. Epub 2019 Jun 4. FASEB J. 2019. PMID: 31162944 Free PMC article.
-
Closing in on human methylation-the versatile family of seven-β-strand (METTL) methyltransferases.Nucleic Acids Res. 2024 Oct 28;52(19):11423-11441. doi: 10.1093/nar/gkae816. Nucleic Acids Res. 2024. PMID: 39351878 Free PMC article. Review.
-
Regulation of eukaryotic elongation factor 1 alpha (eEF1A) by dynamic lysine methylation.RNA Biol. 2018 Mar 4;15(3):314-319. doi: 10.1080/15476286.2018.1440875. Epub 2018 Mar 9. RNA Biol. 2018. PMID: 29447067 Free PMC article.
-
Evolution of Methyltransferase-Like (METTL) Proteins in Metazoa: A Complex Gene Family Involved in Epitranscriptomic Regulation and Other Epigenetic Processes.Mol Biol Evol. 2021 Dec 9;38(12):5309-5327. doi: 10.1093/molbev/msab267. Mol Biol Evol. 2021. PMID: 34480573 Free PMC article.
-
Characterization of the biochemical activity and tumor-promoting role of the dual protein methyltransferase METL-13/METTL13 in Caenorhabditis elegans.PLoS One. 2023 Jun 22;18(6):e0287558. doi: 10.1371/journal.pone.0287558. eCollection 2023. PLoS One. 2023. PMID: 37347777 Free PMC article.
References
-
- Bremang M., Cuomo A., Agresta A. M., Stugiewicz M., Spadotto V., and Bonaldi T. (2013) Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol. Biosyst. 9, 2231–2247 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

