Secretory phospholipase A2 group IIA modulates insulin sensitivity and metabolism
- PMID: 28663239
- PMCID: PMC5580896
- DOI: 10.1194/jlr.M076141
Secretory phospholipase A2 group IIA modulates insulin sensitivity and metabolism
Abstract
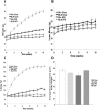
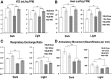
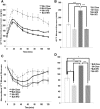
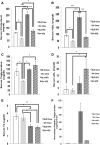
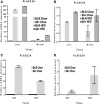
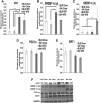
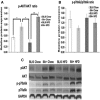
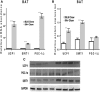
Secretory phospholipase A2 group IIA (PLA2G2A) is a member of a family of secretory phospholipases that have been implicated in inflammation, atherogenesis, and antibacterial actions. Here, we evaluated the role of PLA2G2A in the metabolic response to a high fat diet. C57BL/6 (BL/6) mice do not express PLA2g2a due to a frameshift mutation. We fed BL/6 mice expressing the human PLA2G2A gene (IIA+ mice) a fat diet and assessed the physiologic response. After 10 weeks on the high fat diet, the BL/6 mice were obese, but the IIA+ mice did not gain weight or accumulate lipid. The lean mass in chow- and high fat-fed IIA+ mice was constant and similar to the BL/6 mice on a chow diet. Surprisingly, the IIA+ mice had an elevated metabolic rate, which was not due to differences in physical activity. The IIA+ mice were more insulin sensitive and glucose tolerant than the BL/6 mice, even when the IIA+ mice were provided the high fat diet. The IIA+ mice had increased expression of uncoupling protein 1 (UCP1), sirtuin 1 (SIRT1), and PPARγ coactivator 1α (PGC-1α) in brown adipose tissue (BAT), suggesting that PLA2G2A activates mitochondrial uncoupling in BAT. Our data indicate that PLA2G2A has a previously undiscovered impact on insulin sensitivity and metabolism.
Keywords: hepatic steatosis; high fat diet; insulin resistance; obesity.
Copyright © 2017 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Lambeau G., and Gelb M. H.. 2008. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu. Rev. Biochem. 77: 495–520. - PubMed
-
- Kudo I., and Murakami M.. 2002. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 68–69: 3–58. - PubMed
-
- Murakami M., Taketomi Y., Sato H., and Yamamoto K.. 2011. Secreted phospholipase A2 revisited. J. Biochem. 150: 233–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

