The macrolide antibiotic renaissance
- PMID: 28664582
- PMCID: PMC5573421
- DOI: 10.1111/bph.13936
The macrolide antibiotic renaissance
Abstract
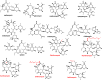
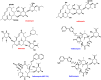
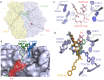
Macrolides represent a large family of protein synthesis inhibitors of great clinical interest due to their applicability to human medicine. Macrolides are composed of a macrocyclic lactone of different ring sizes, to which one or more deoxy-sugar or amino sugar residues are attached. Macrolides act as antibiotics by binding to bacterial 50S ribosomal subunit and interfering with protein synthesis. The high affinity of macrolides for bacterial ribosomes, together with the highly conserved structure of ribosomes across virtually all of the bacterial species, is consistent with their broad-spectrum activity. Since the discovery of the progenitor macrolide, erythromycin, in 1950, many derivatives have been synthesised, leading to compounds with better bioavailability and acid stability and improved pharmacokinetics. These efforts led to the second generation of macrolides, including well-known members such as azithromycin and clarithromycin. Subsequently, in order to address increasing antibiotic resistance, a third generation of macrolides displaying improved activity against many macrolide resistant strains was developed. However, these improvements were accompanied with serious side effects, leading to disappointment and causing many researchers to stop working on macrolide derivatives, assuming that this procedure had reached the end. In contrast, a recent published breakthrough introduced a new chemical platform for synthesis and discovery of a wide range of diverse macrolide antibiotics. This chemical synthesis revolution, in combination with reduction in the side effects, namely, 'Ketek effects', has led to a macrolide renaissance, increasing the hope for novel and safe therapeutic agents to combat serious human infectious diseases.
© 2017 The British Pharmacological Society.
Figures
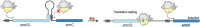
References
-
- Alekshun MN, Levy SB (2007). Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037–1050. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

