Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate
- PMID: 28665412
- PMCID: PMC5497076
- DOI: 10.1038/ncomms16099
Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate
Abstract
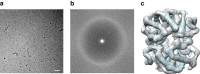
With the advent of direct electron detectors, the perspectives of cryo-electron microscopy (cryo-EM) have changed in a profound way. These cameras are superior to previous detectors in coping with the intrinsically low contrast and beam-induced motion of radiation-sensitive organic materials embedded in amorphous ice, and hence they have enabled the structure determination of many macromolecular assemblies to atomic or near-atomic resolution. Nevertheless, there are still limitations and one of them is the size of the target structure. Here, we report the use of a Volta phase plate in determining the structure of human haemoglobin (64 kDa) at 3.2 Å. Our results demonstrate that this method can be applied to complexes that are significantly smaller than those previously studied by conventional defocus-based approaches. Cryo-EM is now close to becoming a fast and cost-effective alternative to crystallography for high-resolution protein structure determination.
Conflict of interest statement
R.D. is a co-inventor with no financial benefits in US patent US9129774 B2 ‘Method of using a phase plate in a transmission electron microscope’. W.B. is on the Scientific Advisory Board and M.R. an employee of the FEI Company. The remaining author declares no competing financial interests.
Figures

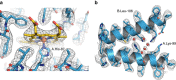
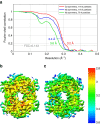
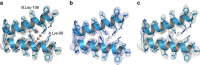
References
-
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 28, 171–193 (1995). - PubMed
-
- Glaeser R. M. Review: electron crystallography: present excitement, a nod to the past, anticipating the future. J. Struct. Biol. 128, 3–14 (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

