Active Vertex Model for cell-resolution description of epithelial tissue mechanics
- PMID: 28665934
- PMCID: PMC5493290
- DOI: 10.1371/journal.pcbi.1005569
Active Vertex Model for cell-resolution description of epithelial tissue mechanics
Abstract
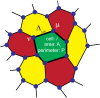
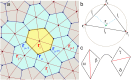
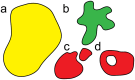
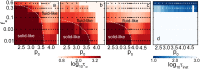
We introduce an Active Vertex Model (AVM) for cell-resolution studies of the mechanics of confluent epithelial tissues consisting of tens of thousands of cells, with a level of detail inaccessible to similar methods. The AVM combines the Vertex Model for confluent epithelial tissues with active matter dynamics. This introduces a natural description of the cell motion and accounts for motion patterns observed on multiple scales. Furthermore, cell contacts are generated dynamically from positions of cell centres. This not only enables efficient numerical implementation, but provides a natural description of the T1 transition events responsible for local tissue rearrangements. The AVM also includes cell alignment, cell-specific mechanical properties, cell growth, division and apoptosis. In addition, the AVM introduces a flexible, dynamically changing boundary of the epithelial sheet allowing for studies of phenomena such as the fingering instability or wound healing. We illustrate these capabilities with a number of case studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



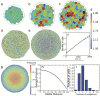

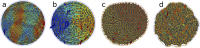
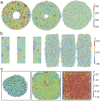
References
-
- Forgacs G, Newman SA. Biological physics of the developing embryo. Cambridge University Press; 2005.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

