Biochemical characterization of functional domains of the chaperone Cosmc
- PMID: 28665962
- PMCID: PMC5493369
- DOI: 10.1371/journal.pone.0180242
Biochemical characterization of functional domains of the chaperone Cosmc
Abstract
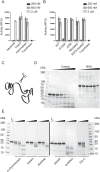
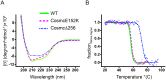
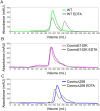
Cosmc is an endoplasmic reticulum chaperone necessary for normal protein O-GalNAc glycosylation through regulation of T-synthase, its single client. Loss-of-function of Cosmc results in expression of the Tn antigen, which is associated with multiple human diseases including cancer. Despite intense interest in dysregulated expression of the Tn antigen, little is known about the structure and function of Cosmc, including domain organization, secondary structure, oligomerization, and co-factors. Limited proteolysis experiments show that Cosmc contains a structured N-terminal domain (CosmcΔ256), and biochemical characterization of CosmcΔ256 reveals wild type chaperone activity. Interestingly, CosmcE152K, which shows loss of function in vivo, exhibits wild type-like activity in vitro. Cosmc and CosmcE152K heterogeneously oligomerize and form monomeric, dimeric, trimeric, and tetrameric species, while CosmcΔ256 is predominantly monomeric as characterized by chemical crosslinking and blue native page electrophoresis. Additionally, Cosmc selectively binds divalent cations in thermal shift assays and metal binding is abrogated by the CosmcΔ256 truncation, and perturbed by the E152K mutation. Therefore, the N-terminal domain of Cosmc mediates T-synthase binding and chaperone function, whereas the C-terminal domain is necessary for oligomerization and metal binding. Our results provide new structure-function insight to Cosmc, indicate that Cosmc behaves as a modular protein and suggests points of modulation or regulation of in vivo chaperone function.
Conflict of interest statement
Figures

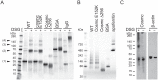
References
-
- Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA. 2002;99(26):16613–8. doi: 10.1073/pnas.262438199 - DOI - PMC - PubMed
-
- Ju T, Cummings RD, Canfield WM. Purification, characterization, and subunit structure of rat core 1 Beta1,3-galactosyltransferase. J Biol Chem. 2002;277(1):169–77. doi: 10.1074/jbc.M109056200 - DOI - PubMed
-
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22(6):736–56. doi: 10.1093/glycob/cwr182 - DOI - PMC - PubMed
-
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KTBG, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–88. doi: 10.1038/emboj.2013.79 - DOI - PMC - PubMed
-
- Ju T, Brewer K, D'Souza A, Cummings RD, Canfield WM. Cloning and expression of human core 1 beta1,3-galactosyltransferase. J Biol Chem. 2002;277(1):178–86. doi: 10.1074/jbc.M109060200 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

