RAS Proteins and Their Regulators in Human Disease
- PMID: 28666118
- PMCID: PMC5555610
- DOI: 10.1016/j.cell.2017.06.009
RAS Proteins and Their Regulators in Human Disease
Abstract
RAS proteins are binary switches, cycling between ON and OFF states during signal transduction. These switches are normally tightly controlled, but in RAS-related diseases, such as cancer, RASopathies, and many psychiatric disorders, mutations in the RAS genes or their regulators render RAS proteins persistently active. The structural basis of the switch and many of the pathways that RAS controls are well known, but the precise mechanisms by which RAS proteins function are less clear. All RAS biology occurs in membranes: a precise understanding of RAS' interaction with membranes is essential to understand RAS action and to intervene in RAS-driven diseases.
Keywords: CRAFT; KRAS; KRAS therapies; NF1; RAF1; RAS effectors; RAS in the membrane; RAS proteins; RAS-driven cancer; RASopathies.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

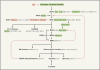



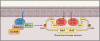

References
-
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 1997;4:686–689. - PubMed
-
- Alvarez-Moya B, López-Alcalá C, Drosten M, Bachs O, Agell N. K-Ras4B phosphorylation at Ser181 is inhibited by calmodulin and modulates K-Ras activity and function. Oncogene. 2010;29:5911–5922. - PubMed
-
- Aoki Y, Niihori T, Inoue S, Matsubara Y. Recent advances in RASopathies. J. Hum. Genet. 2016;61:33–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

