LTR-Retrotransposon Control by tRNA-Derived Small RNAs
- PMID: 28666125
- PMCID: PMC5551035
- DOI: 10.1016/j.cell.2017.06.013
LTR-Retrotransposon Control by tRNA-Derived Small RNAs
Abstract
Transposon reactivation is an inherent danger in cells that lose epigenetic silencing during developmental reprogramming. In the mouse, long terminal repeat (LTR)-retrotransposons, or endogenous retroviruses (ERV), account for most novel insertions and are expressed in the absence of histone H3 lysine 9 trimethylation in preimplantation stem cells. We found abundant 18 nt tRNA-derived small RNA (tRF) in these cells and ubiquitously expressed 22 nt tRFs that include the 3' terminal CCA of mature tRNAs and target the tRNA primer binding site (PBS) essential for ERV reverse transcription. We show that the two most active ERV families, IAP and MusD/ETn, are major targets and are strongly inhibited by tRFs in retrotransposition assays. 22 nt tRFs post-transcriptionally silence coding-competent ERVs, while 18 nt tRFs specifically interfere with reverse transcription and retrotransposon mobility. The PBS offers a unique target to specifically inhibit LTR-retrotransposons, and tRF-targeting is a potentially highly conserved mechanism of small RNA-mediated transposon control.
Keywords: LTR-retrotransposon mobility; epigenetic reprogramming; mouse endogenous retroviruses; small RNA; tRNA fragments; transposon control.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
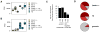


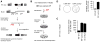

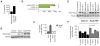

Comment in
-
tsRNAs: new players in mammalian retrotransposon control.Cell Res. 2017 Nov;27(11):1307-1308. doi: 10.1038/cr.2017.109. Epub 2017 Aug 25. Cell Res. 2017. PMID: 28840860 Free PMC article.
References
-
- Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat Genet. 2004;36:534–539. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

