TERRA RNA Antagonizes ATRX and Protects Telomeres
- PMID: 28666128
- PMCID: PMC5552367
- DOI: 10.1016/j.cell.2017.06.017
TERRA RNA Antagonizes ATRX and Protects Telomeres
Abstract
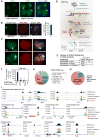
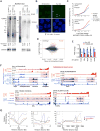
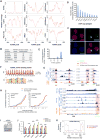
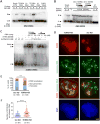
Through an integration of genomic and proteomic approaches to advance understanding of long noncoding RNAs, we investigate the function of the telomeric transcript, TERRA. By identifying thousands of TERRA target sites in the mouse genome, we demonstrate that TERRA can bind both in cis to telomeres and in trans to genic targets. We then define a large network of interacting proteins, including epigenetic factors, telomeric proteins, and the RNA helicase, ATRX. TERRA and ATRX share hundreds of target genes and are functionally antagonistic at these loci: whereas TERRA activates, ATRX represses gene expression. At telomeres, TERRA competes with telomeric DNA for ATRX binding, suppresses ATRX localization, and ensures telomeric stability. Depleting TERRA increases telomerase activity and induces telomeric pathologies, including formation of telomere-induced DNA damage foci and loss or duplication of telomeric sequences. We conclude that TERRA functions as an epigenomic modulator in trans and as an essential regulator of telomeres in cis.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Non-coding RNA: A protective role for TERRA at telomeres.Nat Rev Genet. 2017 Jul 17;18(8):453. doi: 10.1038/nrg.2017.58. Nat Rev Genet. 2017. PMID: 28713153 No abstract available.
References
-
- Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. - PubMed
-
- Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25:29–36. - PubMed
-
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science. 2007;318:798–801. - PubMed
-
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–1205. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

