The Structured Coalescent and Its Approximations
- PMID: 28666382
- PMCID: PMC5850743
- DOI: 10.1093/molbev/msx186
The Structured Coalescent and Its Approximations
Abstract
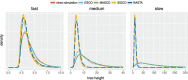
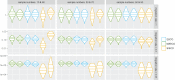
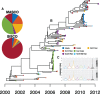
Phylogeographic methods can help reveal the movement of genes between populations of organisms. This has been widely done to quantify pathogen movement between different host populations, the migration history of humans, and the geographic spread of languages or gene flow between species using the location or state of samples alongside sequence data. Phylogenies therefore offer insights into migration processes not available from classic epidemiological or occurrence data alone. Phylogeographic methods have however several known shortcomings. In particular, one of the most widely used methods treats migration the same as mutation, and therefore does not incorporate information about population demography. This may lead to severe biases in estimated migration rates for data sets where sampling is biased across populations. The structured coalescent on the other hand allows us to coherently model the migration and coalescent process, but current implementations struggle with complex data sets due to the need to infer ancestral migration histories. Thus, approximations to the structured coalescent, which integrate over all ancestral migration histories, have been developed. However, the validity and robustness of these approximations remain unclear. We present an exact numerical solution to the structured coalescent that does not require the inference of migration histories. Although this solution is computationally unfeasible for large data sets, it clarifies the assumptions of previously developed approximate methods and allows us to provide an improved approximation to the structured coalescent. We have implemented these methods in BEAST2, and we show how these methods compare under different scenarios.
Keywords: infectious diseases; migration; phylodynamics; phylogenetics; phylogeography; population structure.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




Similar articles
-
New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation.PLoS Genet. 2015 Aug 12;11(8):e1005421. doi: 10.1371/journal.pgen.1005421. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26267488 Free PMC article.
-
Bayesian Inference of Pathogen Phylogeography using the Structured Coalescent Model.PLoS Comput Biol. 2025 Apr 21;21(4):e1012995. doi: 10.1371/journal.pcbi.1012995. eCollection 2025 Apr. PLoS Comput Biol. 2025. PMID: 40258093 Free PMC article.
-
Recombination-aware phylogeographic inference using the structured coalescent with ancestral recombination.PLoS Comput Biol. 2022 Aug 19;18(8):e1010422. doi: 10.1371/journal.pcbi.1010422. eCollection 2022 Aug. PLoS Comput Biol. 2022. PMID: 35984849 Free PMC article.
-
Phylogeographic Approaches to Characterize the Emergence of Plant Pathogens.Phytopathology. 2021 Jan;111(1):68-77. doi: 10.1094/PHYTO-07-20-0319-FI. Epub 2020 Nov 21. Phytopathology. 2021. PMID: 33021879 Review.
-
Ancestral Population Genomics.Methods Mol Biol. 2019;1910:555-589. doi: 10.1007/978-1-4939-9074-0_18. Methods Mol Biol. 2019. PMID: 31278677 Review.
Cited by
-
nosoi: A stochastic agent-based transmission chain simulation framework in r.Methods Ecol Evol. 2020 Aug;11(8):1002-1007. doi: 10.1111/2041-210X.13422. Epub 2020 Jun 21. Methods Ecol Evol. 2020. PMID: 32983401 Free PMC article.
-
Multiclonal human origin and global expansion of an endemic bacterial pathogen of livestock.Proc Natl Acad Sci U S A. 2022 Dec 13;119(50):e2211217119. doi: 10.1073/pnas.2211217119. Epub 2022 Dec 5. Proc Natl Acad Sci U S A. 2022. PMID: 36469788 Free PMC article.
-
Evolutionary evidence for multi-host transmission of cetacean morbillivirus.Emerg Microbes Infect. 2018 Dec 5;7(1):201. doi: 10.1038/s41426-018-0207-x. Emerg Microbes Infect. 2018. PMID: 30514855 Free PMC article.
-
Inferring time-dependent migration and coalescence patterns from genetic sequence and predictor data in structured populations.Virus Evol. 2019 Aug 14;5(2):vez030. doi: 10.1093/ve/vez030. eCollection 2019 Jul. Virus Evol. 2019. PMID: 31428459 Free PMC article.
-
MASCOT: parameter and state inference under the marginal structured coalescent approximation.Bioinformatics. 2018 Nov 15;34(22):3843-3848. doi: 10.1093/bioinformatics/bty406. Bioinformatics. 2018. PMID: 29790921 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

