RGIFE: a ranked guided iterative feature elimination heuristic for the identification of biomarkers
- PMID: 28666416
- PMCID: PMC5493069
- DOI: 10.1186/s12859-017-1729-2
RGIFE: a ranked guided iterative feature elimination heuristic for the identification of biomarkers
Abstract
Background: Current -omics technologies are able to sense the state of a biological sample in a very wide variety of ways. Given the high dimensionality that typically characterises these data, relevant knowledge is often hidden and hard to identify. Machine learning methods, and particularly feature selection algorithms, have proven very effective over the years at identifying small but relevant subsets of variables from a variety of application domains, including -omics data. Many methods exist with varying trade-off between the size of the identified variable subsets and the predictive power of such subsets. In this paper we focus on an heuristic for the identification of biomarkers called RGIFE: Rank Guided Iterative Feature Elimination. RGIFE is guided in its biomarker identification process by the information extracted from machine learning models and incorporates several mechanisms to ensure that it creates minimal and highly predictive features sets.
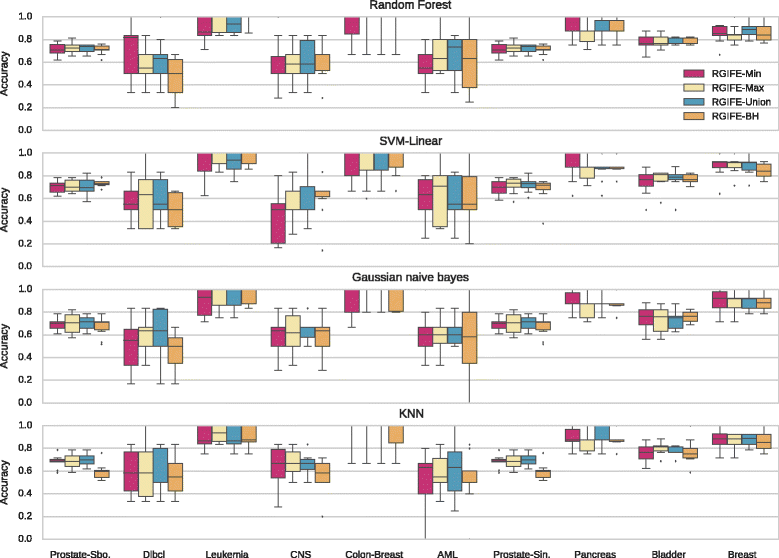
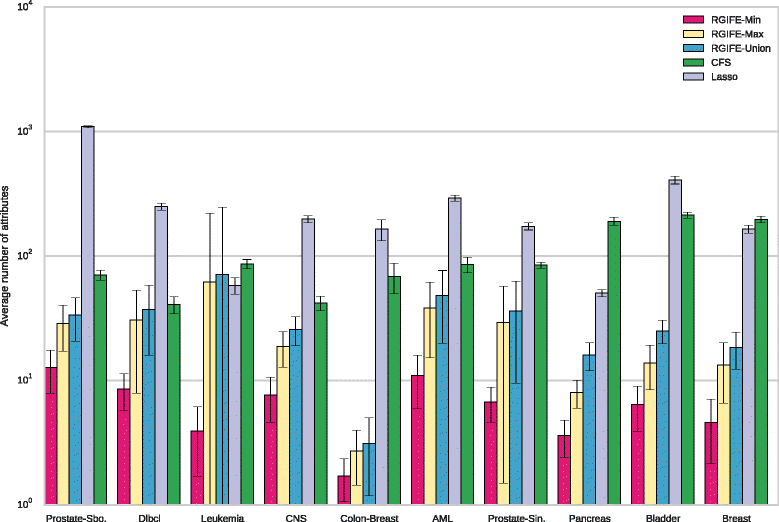
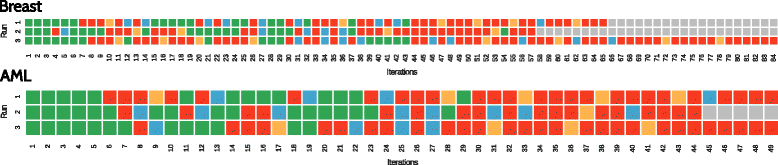
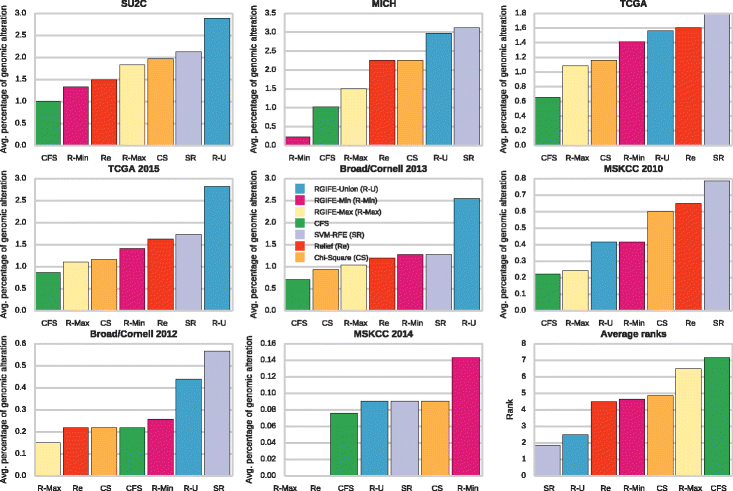
Results: We compare RGIFE against five well-known feature selection algorithms using both synthetic and real (cancer-related transcriptomics) datasets. First, we assess the ability of the methods to identify relevant and highly predictive features. Then, using a prostate cancer dataset as a case study, we look at the biological relevance of the identified biomarkers.
Conclusions: We propose RGIFE, a heuristic for the inference of reduced panels of biomarkers that obtains similar predictive performance to widely adopted feature selection methods while selecting significantly fewer feature. Furthermore, focusing on the case study, we show the higher biological relevance of the biomarkers selected by our approach. The RGIFE source code is available at: http://ico2s.org/software/rgife.html .
Keywords: Biomarkers; Feature reduction; Knowledge extraction; Machine learning.
Figures


References
-
- Inza IN, Calvo B, Armañanzas R, Bengoetxea E, Larrañaga P, Lozano J. Bioinformatics Methods in Clinical Research. Methods in Molecular Biology. Springer: Humana Press; 2010. Machine learning: An indispensable tool in bioinformatics. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

