Untangling Genetic Risk for Alzheimer's Disease
- PMID: 28666525
- PMCID: PMC5699970
- DOI: 10.1016/j.biopsych.2017.05.014
Untangling Genetic Risk for Alzheimer's Disease
Abstract
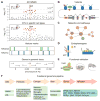
Alzheimer's disease (AD) is a genetically heterogeneous neurodegenerative disorder caused by fully penetrant single gene mutations in a minority of cases, while the majority of cases are sporadic or show modest familial clustering. These cases are of late onset and likely result from the interaction of many genes and the environment. More than 30 loci have been implicated in AD by a combination of linkage, genome-wide association, and whole genome/exome sequencing. We have learned from these studies that perturbations in endolysosomal, lipid metabolism, and immune response pathways substantially contribute to sporadic AD pathogenesis. We review here current knowledge about functions of AD susceptibility genes, highlighting cells of the myeloid lineage as drivers of at least part of the genetic component in late-onset AD. Although targeted resequencing utilized for the identification of causal variants has discovered coding mutations in some AD-associated genes, a lot of risk variants lie in noncoding regions. Here we discuss the use of functional genomics approaches that integrate transcriptomic, epigenetic, and endophenotype traits with systems biology to annotate genetic variants, and to facilitate discovery of AD risk genes. Further validation in cell culture and mouse models will be necessary to establish causality for these genes. This knowledge will allow mechanism-based design of novel therapeutic interventions in AD and promises coherent implementation of treatment in a personalized manner.
Keywords: Alzheimer’s disease; Endolysosomal pathway; Functional genomics; Genome-wide association studies; Immune response; Lipid metabolism.
Copyright © 2017 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

