Initial Gut Microbial Composition as a Key Factor Driving Host Response to Antibiotic Treatment, as Exemplified by the Presence or Absence of Commensal Escherichia coli
- PMID: 28667114
- PMCID: PMC5561281
- DOI: 10.1128/AEM.01107-17
Initial Gut Microbial Composition as a Key Factor Driving Host Response to Antibiotic Treatment, as Exemplified by the Presence or Absence of Commensal Escherichia coli
Abstract
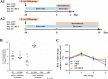
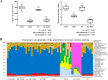
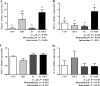
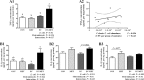
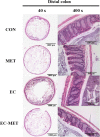
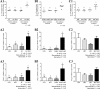
Antibiotics are important for treating bacterial infection; however, efficacies and side effects of antibiotics vary in medicine and experimental models. A few studies have correlated microbiota composition variations with health outcomes in response to antibiotics; however, no study has demonstrated causality. We had noted variation in colonic expression of C-type lectins, regenerating islet-derived protein 3β (Reg3β) and Reg3γ, after metronidazole treatment in a mouse model. To investigate the effects of specific variations in the preexisting microbiome on host response to antibiotics, mice harboring a normal microbiota were allocated to 4 treatments in a 2-by-2 factorial arrangement with or without commensal Escherichia coli and with or without metronidazole in drinking water. E. coli colonized readily without causing a notable shift in the microbiota or host response. Metronidazole administration reduced microbiota biodiversity, indicated by decreased Chao1 and Shannon index values, and altered microbiota composition. However, the presence of E. coli strongly affected metronidazole-induced microbiota shifts. Remarkably, this single commensal bacterium in the context of a complex population led to variations in host responses to metronidazole treatment, including increased expression of antimicrobial peptides Reg3β and Reg3γ and intestinal inflammation indicated by tumor necrosis factor alpha levels. Similar results were obtained from 2-week antibiotic exposure and with additional E. coli isolates. The results of this proof-of-concept study indicate that even minor variations in initial commensal microbiota can drive shifts in microbial composition and host response after antibiotic administration. As well as providing an explanation for variability in animal models using antibiotics, the findings encourage the development of personalized medication in antibiotic therapies.IMPORTANCE This work provides an understanding of variability in studies where antibiotics are used to alter the gut microbiota to generate a host response. Furthermore, although providing evidence only for the one antibiotic, the study demonstrated that initial gut microbial composition is a key factor driving host response to antibiotic administration, creating a compelling argument for considering personalized medication based on individual variations in gut microbiota.
Keywords: Escherichia coli; gut microbiota; metronidazole; regenerating islet-derived protein 3β (Reg3β); regenerating islet-derived protein 3γ (Reg3γ).
Copyright © 2017 American Society for Microbiology.
Figures

Similar articles
-
A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota.Gut Microbes. 2018;9(5):391-399. doi: 10.1080/19490976.2018.1447291. Epub 2018 Aug 24. Gut Microbes. 2018. PMID: 29517960 Free PMC article.
-
The effect of antibiotic cocktails on host immune status is dynamic and does not always correspond to changes in gut microbiota.Appl Microbiol Biotechnol. 2020 Jun;104(11):4995-5009. doi: 10.1007/s00253-020-10611-1. Epub 2020 Apr 18. Appl Microbiol Biotechnol. 2020. PMID: 32303819
-
Comparison of different modes of antibiotic delivery on gut microbiota depletion efficiency and body composition in mouse.BMC Microbiol. 2020 Nov 11;20(1):340. doi: 10.1186/s12866-020-02018-9. BMC Microbiol. 2020. PMID: 33176677 Free PMC article.
-
Role of antibiotics for treatment of inflammatory bowel disease.World J Gastroenterol. 2016 Jan 21;22(3):1078-87. doi: 10.3748/wjg.v22.i3.1078. World J Gastroenterol. 2016. PMID: 26811648 Free PMC article. Review.
-
Disruption of the Gut Ecosystem by Antibiotics.Yonsei Med J. 2018 Jan;59(1):4-12. doi: 10.3349/ymj.2018.59.1.4. Yonsei Med J. 2018. PMID: 29214770 Free PMC article. Review.
Cited by
-
Antibiotic associated diarrhea in outpatient pediatric antibiotic therapy.BMC Pediatr. 2023 Mar 18;23(1):121. doi: 10.1186/s12887-023-03939-w. BMC Pediatr. 2023. PMID: 36932373 Free PMC article.
-
Association between in-hospital antibiotic use and long-term outcomes in critically ill patients.Antimicrob Steward Healthc Epidemiol. 2025 Jun 23;5(1):e135. doi: 10.1017/ash.2025.10054. eCollection 2025. Antimicrob Steward Healthc Epidemiol. 2025. PMID: 40567396 Free PMC article.
-
Isolation and Characterization of a Novel Temperate Escherichia coli Bacteriophage, Kapi1, Which Modifies the O-Antigen and Contributes to the Competitiveness of Its Host during Colonization of the Murine Gastrointestinal Tract.mBio. 2022 Feb 22;13(1):e0208521. doi: 10.1128/mbio.02085-21. Epub 2022 Jan 25. mBio. 2022. PMID: 35073745 Free PMC article.
-
Isolation of Commensal Escherichia coli Strains from Feces of Healthy Laboratory Mice or Rats.Bio Protoc. 2018 Mar 20;8(6):e2780. doi: 10.21769/BioProtoc.2780. eCollection 2018 Mar 20. Bio Protoc. 2018. PMID: 34179293 Free PMC article.
-
Microbial Fermentation of Dietary Protein: An Important Factor in Diet⁻Microbe⁻Host Interaction.Microorganisms. 2019 Jan 13;7(1):19. doi: 10.3390/microorganisms7010019. Microorganisms. 2019. PMID: 30642098 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

