Population genetic structure in Sabatieria (Nematoda) reveals intermediary gene flow and admixture between distant cold seeps from the Mediterranean Sea
- PMID: 28668078
- PMCID: PMC5494145
- DOI: 10.1186/s12862-017-1003-2
Population genetic structure in Sabatieria (Nematoda) reveals intermediary gene flow and admixture between distant cold seeps from the Mediterranean Sea
Abstract
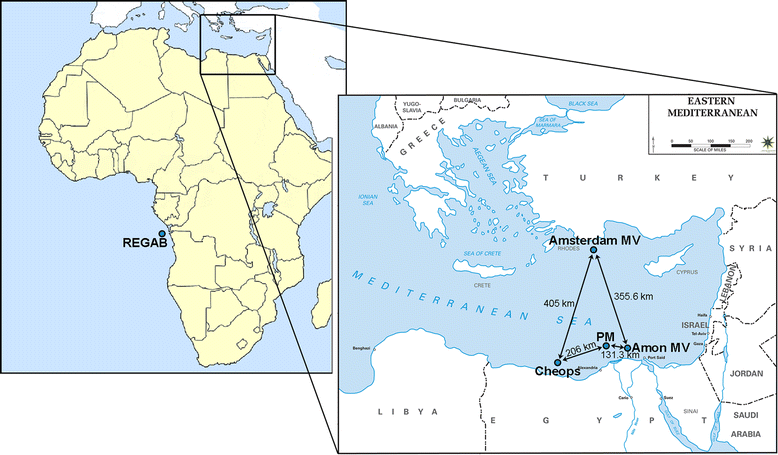
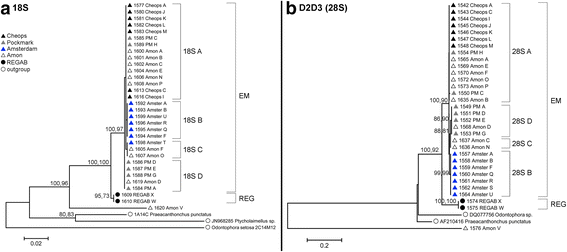
Background: There is a general lack of information on the dispersal and genetic structuring for populations of small-sized deep-water taxa, including free-living nematodes which inhabit and dominate the seafloor sediments. This is also true for unique and scattered deep-sea habitats such as cold seeps. Given the limited dispersal capacity of marine nematodes, genetic differentiation between such geographically isolated habitat patches is expected to be high. Against this background, we examined genetic variation in both mitochondrial (COI) and nuclear (18S and 28S ribosomal) DNA markers of 333 individuals of the genus Sabatieria, abundantly present in reduced cold-seep sediments. Samples originated from four Eastern Mediterranean cold seeps, separated by hundreds of kilometers, and one seep in the Southeast Atlantic.
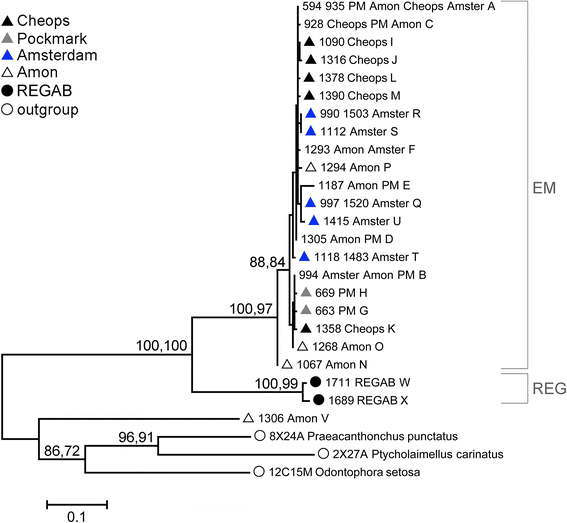
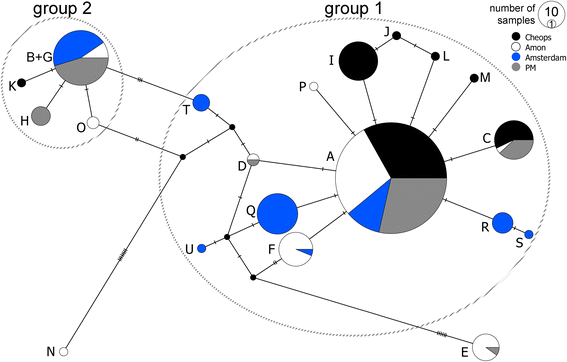
Results: Individuals from the Mediterranean and Atlantic were divided into two separate but closely-related species clades. Within the Eastern Mediterranean, all specimens belonged to a single species, but with a strong population genetic structure (ΦST = 0.149). The haplotype network of COI contained 19 haplotypes with the most abundant haplotype (52% of the specimens) shared between all four seeps. The number of private haplotypes was high (15), but the number of mutations between haplotypes was low (1-8). These results indicate intermediary gene flow among the Mediterranean Sabatieria populations with no evidence of long-term barriers to gene flow.
Conclusions: The presence of shared haplotypes and multiple admixture events indicate that Sabatieria populations from disjunct cold seeps are not completely isolated, with gene flow most likely facilitated through water current transportation of individuals and/or eggs. Genetic structure and molecular diversity indices are comparable to those of epiphytic shallow-water marine nematodes, while no evidence of sympatric cryptic species was found for the cold-seep Sabatieria.
Conflict of interest statement
Ethics approval and consent to participate
The manuscript does not report on or involve the use of any vertebrate or human data or tissue, therefore this section is not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Distinct genetic differentiation and species diversification within two marine nematodes with different habitat preference in Antarctic sediments.BMC Evol Biol. 2017 May 30;17(1):120. doi: 10.1186/s12862-017-0968-1. BMC Evol Biol. 2017. PMID: 28558672 Free PMC article.
-
Hydroids (Cnidaria, Hydrozoa) from Mauritanian Coral Mounds.Zootaxa. 2020 Nov 16;4878(3):zootaxa.4878.3.2. doi: 10.11646/zootaxa.4878.3.2. Zootaxa. 2020. PMID: 33311142
-
Low genetic but high morphological variation over more than 1000 km coastline refutes omnipresence of cryptic diversity in marine nematodes.BMC Evol Biol. 2017 Mar 7;17(1):71. doi: 10.1186/s12862-017-0908-0. BMC Evol Biol. 2017. PMID: 28270090 Free PMC article.
-
Ecology and biogeography of free-living nematodes associated with chemosynthetic environments in the deep sea: a review.PLoS One. 2010 Aug 27;5(8):e12449. doi: 10.1371/journal.pone.0012449. PLoS One. 2010. PMID: 20805986 Free PMC article. Review.
-
Pillars of Hercules: is the Atlantic-Mediterranean transition a phylogeographical break?Mol Ecol. 2007 Nov;16(21):4426-44. doi: 10.1111/j.1365-294X.2007.03477.x. Epub 2007 Oct 1. Mol Ecol. 2007. PMID: 17908222 Review.
Cited by
-
First molecular observation on Mylonchulus hawaiiensis from South Africa.Helminthologia. 2024 Apr 23;61(1):99-108. doi: 10.2478/helm-2024-0010. eCollection 2024 Mar. Helminthologia. 2024. PMID: 38659467 Free PMC article.
-
New and known free-living nematode species (Nematoda: Chromadorea) from offshore tsunami monitoring buoys in the Southwest Pacific Ocean.PeerJ. 2025 Jul 30;13:e19789. doi: 10.7717/peerj.19789. eCollection 2025. PeerJ. 2025. PMID: 40755792 Free PMC article.
References
-
- Gage JD, Tyler PA. Deep-Sea biology: a natural history of organisms at the Deep-Sea floor. Cambridge, UK: Cambridge University Press; 1991. p. 504.
-
- Brandt A, Blazewicz-Paszkowycz M, Bamber RN, Mühlenhardt-Siegel U, Malyutina MV, Kaiser S, et al. Are there widespread pericarid species in the deep sea (Crustacea: malacostraca)? Polish Polar Research. 2012;33(2):139–62. doi:10.2478/v10183-012-0012-5.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

