Ultra-sensitive Sequencing Identifies High Prevalence of Clonal Hematopoiesis-Associated Mutations throughout Adult Life
- PMID: 28669404
- PMCID: PMC5501773
- DOI: 10.1016/j.ajhg.2017.05.013
Ultra-sensitive Sequencing Identifies High Prevalence of Clonal Hematopoiesis-Associated Mutations throughout Adult Life
Abstract
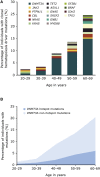
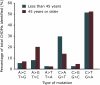
Clonal hematopoiesis results from somatic mutations in hematopoietic stem cells, which give an advantage to mutant cells, driving their clonal expansion and potentially leading to leukemia. The acquisition of clonal hematopoiesis-driver mutations (CHDMs) occurs with normal aging and these mutations have been detected in more than 10% of individuals ≥65 years. We aimed to examine the prevalence and characteristics of CHDMs throughout adult life. We developed a targeted re-sequencing assay combining high-throughput with ultra-high sensitivity based on single-molecule molecular inversion probes (smMIPs). Using smMIPs, we screened more than 100 loci for CHDMs in more than 2,000 blood DNA samples from population controls between 20 and 69 years of age. Loci screened included 40 regions known to drive clonal hematopoiesis when mutated and 64 novel candidate loci. We identified 224 somatic mutations throughout our cohort, of which 216 were coding mutations in known driver genes (DNMT3A, JAK2, GNAS, TET2, and ASXL1), including 196 point mutations and 20 indels. Our assay's improved sensitivity allowed us to detect mutations with variant allele frequencies as low as 0.001. CHDMs were identified in more than 20% of individuals 60 to 69 years of age and in 3% of individuals 20 to 29 years of age, approximately double the previously reported prevalence despite screening a limited set of loci. Our findings support the occurrence of clonal hematopoiesis-associated mutations as a widespread mechanism linked with aging, suggesting that mosaicism as a result of clonal evolution of cells harboring somatic mutations is a universal mechanism occurring at all ages in healthy humans.
Keywords: DNMT3A; age-related clonal hematopoiesis; mosaicism; smMIPs; somatic mutations; ultra-sensitive sequencing.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- McKerrell T., Park N., Moreno T., Grove C.S.S., Ponstingl H., Stephens J., Crawley C., Craig J., Scott M.A.A., Hodkinson C., Understanding Society Scientific Group Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–1245. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

