Development of a Novel Lead that Targets M. tuberculosis Polyketide Synthase 13
- PMID: 28669536
- PMCID: PMC5509550
- DOI: 10.1016/j.cell.2017.06.025
Development of a Novel Lead that Targets M. tuberculosis Polyketide Synthase 13
Abstract
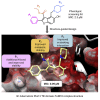
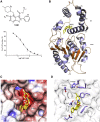
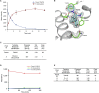
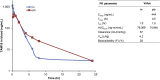
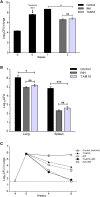
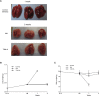
Widespread resistance to first-line TB drugs is a major problem that will likely only be resolved through the development of new drugs with novel mechanisms of action. We have used structure-guided methods to develop a lead molecule that targets the thioesterase activity of polyketide synthase Pks13, an essential enzyme that forms mycolic acids, required for the cell wall of Mycobacterium tuberculosis. Our lead, TAM16, is a benzofuran class inhibitor of Pks13 with highly potent in vitro bactericidal activity against drug-susceptible and drug-resistant clinical isolates of M. tuberculosis. In multiple mouse models of TB infection, TAM16 showed in vivo efficacy equal to the first-line TB drug isoniazid, both as a monotherapy and in combination therapy with rifampicin. TAM16 has excellent pharmacological and safety profiles, and the frequency of resistance for TAM16 is ∼100-fold lower than INH, suggesting that it can be developed as a new antitubercular aimed at the acute infection. PAPERCLIP.
Keywords: Mycobacterium tuberculosis; Pks13 thioesterase domain; benzofuran inhibitors; crystal structure; polyketide synthase; structure-based drug discovery.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

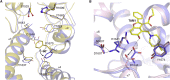
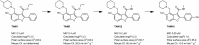
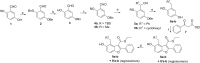
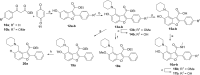


Comment in
-
Antibacterial agents: New routes to tuberculosis treatment.Nat Rev Drug Discov. 2017 Sep;16(9):600-601. doi: 10.1038/nrd.2017.156. Epub 2017 Aug 11. Nat Rev Drug Discov. 2017. PMID: 28799546 No abstract available.
References
-
- Alelyunas Y.W., Liu R., Pelosi-Kilby L., Shen C. Application of a Dried-DMSO rapid throughput 24-h equilibrium solubility in advancing discovery candidates. Eur. J. Pharm. Sci. 2009;37:172–182. - PubMed
-
- Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K.S., Wilson T., Collins D., de Lisle G., Jacobs W.R., Jr. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. - PubMed
-
- Barrero A.F., del Moral J.F.Q., Herrador M.M., Arteaga P., Cortés M., Benites J., Rosellón A. Mild and rapid method for the generation of o-quinone methide intermediates. Synthesis of puupehedione analogues. Tetrahedron. 2006;62:6012–6017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

