Complement Evasion Strategies of Viruses: An Overview
- PMID: 28670306
- PMCID: PMC5472698
- DOI: 10.3389/fmicb.2017.01117
Complement Evasion Strategies of Viruses: An Overview
Abstract
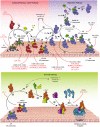
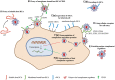
Being a major first line of immune defense, the complement system keeps a constant vigil against viruses. Its ability to recognize large panoply of viruses and virus-infected cells, and trigger the effector pathways, results in neutralization of viruses and killing of the infected cells. This selection pressure exerted by complement on viruses has made them evolve a multitude of countermeasures. These include targeting the recognition molecules for the avoidance of detection, targeting key enzymes and complexes of the complement pathways like C3 convertases and C5b-9 formation - either by encoding complement regulators or by recruiting membrane-bound and soluble host complement regulators, cleaving complement proteins by encoding protease, and inhibiting the synthesis of complement proteins. Additionally, viruses also exploit the complement system for their own benefit. For example, they use complement receptors as well as membrane regulators for cellular entry as well as their spread. Here, we provide an overview on the complement subversion mechanisms adopted by the members of various viral families including Poxviridae, Herpesviridae, Adenoviridae, Flaviviridae, Retroviridae, Picornaviridae, Astroviridae, Togaviridae, Orthomyxoviridae and Paramyxoviridae.
Keywords: DNA viruses; RNA viruses; complement; complement evasion; immune evasion; innate immunity; pathogenesis; retroviruses.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

