Ancient selection for derived alleles at a GDF5 enhancer influencing human growth and osteoarthritis risk
- PMID: 28671685
- PMCID: PMC6556117
- DOI: 10.1038/ng.3911
Ancient selection for derived alleles at a GDF5 enhancer influencing human growth and osteoarthritis risk
Abstract
Variants in GDF5 are associated with human arthritis and decreased height, but the causal mutations are still unknown. We surveyed the Gdf5 locus for regulatory regions in transgenic mice and fine-mapped separate enhancers controlling expression in joints versus growing ends of long bones. A large downstream regulatory region contains a novel growth enhancer (GROW1), which is required for normal Gdf5 expression at ends of developing bones and for normal bone lengths in vivo. Human GROW1 contains a common base-pair change that decreases enhancer activity and colocalizes with peaks of positive selection in humans. The derived allele is rare in Africa but common in Eurasia and is found in Neandertals and Denisovans. Our results suggest that an ancient regulatory variant in GROW1 has been repeatedly selected in northern environments and that past selection on growth phenotypes explains the high frequency of a GDF5 haplotype that also increases arthritis susceptibility in many human populations.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing financial interests.
Figures



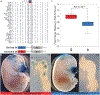
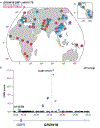


References
-
- Miyamoto Y et al. A functional polymorphism in the 5’UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet 39, 529–33 (2007). - PubMed
-
- Zhang R et al. A comprehensive meta-analysis of association between genetic variants of GDF5 and osteoarthritis of the knee, hip and hand. Inflamm Res 64, 405–14 (2015). - PubMed
METHODS-ONLY REFERENCES
-
- Iafrate AJ et al. Detection of large-scale variation in the human genome. Nat Genet 36, 949–51 (2004). - PubMed
-
- Settle SH et al. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol 254, 116–30 (2003). - PubMed
-
- Yang L, Mali P, Kim-Kiselak C & Church G CRISPR-Cas-mediated targeted genome editing in human cells. Methods Mol Biol 1114, 245–67 (2014). - PubMed
-
- McLeod MJ Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red. S. Teratology 22, 299–301 (1980). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

