Distinct cis elements in the 3' UTR of the C. elegans cebp-1 mRNA mediate its regulation in neuronal development
- PMID: 28673818
- PMCID: PMC5828015
- DOI: 10.1016/j.ydbio.2017.06.022
Distinct cis elements in the 3' UTR of the C. elegans cebp-1 mRNA mediate its regulation in neuronal development
Abstract
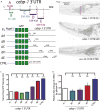
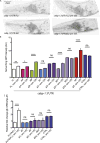
The 3' untranslated regions (3' UTRs) of mRNAs mediate post-transcriptional regulation of genes in many biological processes. Cis elements in 3' UTRs can interact with RNA-binding factors in sequence-specific or structure-dependent manners, enabling regulation of mRNA stability, translation, and localization. Caenorhabditis elegans CEBP-1 is a conserved transcription factor of the C/EBP family, and functions in diverse contexts, from neuronal development and axon regeneration to organismal growth. Previous studies revealed that the levels of cebp-1 mRNA in neurons depend on its 3' UTR and are also negatively regulated by the E3 ubiquitin ligase RPM-1. Here, by systematically dissecting cebp-1's 3' UTR, we test the roles of specific cis elements in cebp-1 expression and function in neurons. We present evidence for a putative stem-loop in the cebp-1 3' UTR that contributes to basal expression levels of mRNA and to negative regulation by rpm-1. Mutant animals lacking the endogenous cebp-1 3' UTR showed a noticeable increased expression of cebp-1 mRNA and enhanced the neuronal developmental phenotypes of rpm-1 mutants. Our data reveal multiple cis elements within cebp-1's 3' UTR that help to optimize CEBP-1 expression levels in neuronal development.
Keywords: Axon regeneration; Mechanosensory neurons; Stem-loop; Translation regulation; mRNA secondary structure; mRNA stability; rpm-1.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
A conserved sequence motif in 3' untranslated regions of ribosomal protein mRNAs in nematodes.RNA. 2006 Oct;12(10):1786-9. doi: 10.1261/rna.51306. Epub 2006 Aug 17. RNA. 2006. PMID: 16917125 Free PMC article.
-
Coordinated inhibition of C/EBP by Tribbles in multiple tissues is essential for Caenorhabditis elegans development.BMC Biol. 2016 Dec 7;14(1):104. doi: 10.1186/s12915-016-0320-z. BMC Biol. 2016. PMID: 27927209 Free PMC article.
-
Repression by the 3' UTR of fem-3, a sex-determining gene, relies on a ubiquitous mog-dependent control in Caenorhabditis elegans.EMBO J. 1998 Nov 2;17(21):6337-47. doi: 10.1093/emboj/17.21.6337. EMBO J. 1998. PMID: 9799241 Free PMC article.
-
Emerging Roles for 3' UTRs in Neurons.Int J Mol Sci. 2020 May 12;21(10):3413. doi: 10.3390/ijms21103413. Int J Mol Sci. 2020. PMID: 32408514 Free PMC article. Review.
-
Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression.Bioessays. 2002 Feb;24(2):119-29. doi: 10.1002/bies.10046. Bioessays. 2002. PMID: 11835276 Review.
Cited by
-
Functional Dissection of C. elegans bZip-Protein CEBP-1 Reveals Novel Structural Motifs Required for Axon Regeneration and Nuclear Import.Front Cell Neurosci. 2019 Jul 31;13:348. doi: 10.3389/fncel.2019.00348. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31417366 Free PMC article.
-
The kpc-1 3'UTR facilitates dendritic transport and translation efficiency of mRNAs for dendrite arborization of a mechanosensory neuron important for male courtship.PLoS Genet. 2024 Aug 7;20(8):e1011362. doi: 10.1371/journal.pgen.1011362. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39110773 Free PMC article.
-
Methylmercury Induces Metabolic Alterations in Caenorhabditis elegans: Role for C/EBP Transcription Factor.Toxicol Sci. 2020 Mar 1;174(1):112-123. doi: 10.1093/toxsci/kfz244. Toxicol Sci. 2020. PMID: 31851340 Free PMC article.
-
The role of structure in regulatory RNA elements.Biosci Rep. 2024 Oct 30;44(10):BSR20240139. doi: 10.1042/BSR20240139. Biosci Rep. 2024. PMID: 39364891 Free PMC article. Review.
-
Neurogenesis in Caenorhabditis elegans.Genetics. 2024 Oct 7;228(2):iyae116. doi: 10.1093/genetics/iyae116. Genetics. 2024. PMID: 39167071 Free PMC article. Review.
References
-
- Belasco JG, Chen CY. Mechanism of puf mRNA degradation: the role of an intercistronic stem-loop structure. Gene. 1988;72:109–117. - PubMed
-
- Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9:333–336. - PubMed
-
- Chen CY, Beatty JT, Cohen SN, Belasco JG. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988;52:609–619. - PubMed
-
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA Translation and Stability for microRNAs. Annu Rev Biochem. 2010;79:351–379. - PubMed
-
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

