Yeast RNA-Binding Protein Nab3 Regulates Genes Involved in Nitrogen Metabolism
- PMID: 28674185
- PMCID: PMC5574042
- DOI: 10.1128/MCB.00154-17
Yeast RNA-Binding Protein Nab3 Regulates Genes Involved in Nitrogen Metabolism
Abstract
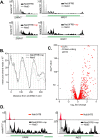
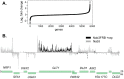
Termination of Saccharomyces cerevisiae RNA polymerase II (Pol II) transcripts occurs through two alternative pathways. Termination of mRNAs is coupled to cleavage and polyadenylation while noncoding transcripts are terminated through the Nrd1-Nab3-Sen1 (NNS) pathway in a process that is linked to RNA degradation by the nuclear exosome. Some mRNA transcripts are also attenuated through premature termination directed by the NNS complex. In this paper we present the results of nuclear depletion of the NNS component Nab3. As expected, many noncoding RNAs fail to terminate properly. In addition, we observe that nitrogen catabolite-repressed genes are upregulated by Nab3 depletion.
Keywords: nitrogen metabolism; noncoding RNA; termination; transcription.
Copyright © 2017 American Society for Microbiology.
Figures


References
-
- Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A. 2010. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol 11:R87. doi:10.1186/gb-2010-11-8-r87. - DOI - PMC - PubMed
-
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725–737. doi:10.1016/j.cell.2005.04.030. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
