Two proteases with caspase-3-like activity, cathepsin B and proteasome, antagonistically control ER-stress-induced programmed cell death in Arabidopsis
- PMID: 28675441
- PMCID: PMC5947621
- DOI: 10.1111/nph.14676
Two proteases with caspase-3-like activity, cathepsin B and proteasome, antagonistically control ER-stress-induced programmed cell death in Arabidopsis
Abstract
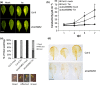
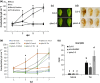
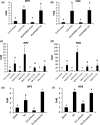
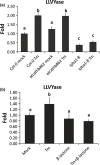
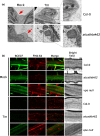
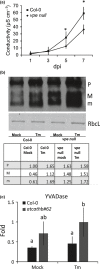
Programmed cell death (PCD) induced by endoplasmic reticulum (ER) stress is implicated in various plant physiological processes, yet its mechanism is still elusive. An activation of caspase-3-like enzymatic activity was clearly demonstrated but the role of the two known plant proteases with caspase-3-like activity, cathepsin B and proteasome subunit PBA1, remains to be established. Both genetic downregulation and chemical inhibition were used to investigate the function of cathepsin B and PBA1 in ER-stress-induced PCD (ERSID). Transcript level and activity labelling of cathepsin B were used to assess activation. To study tonoplast rupture, a plant PCD feature, both confocal and electronic microscopies were used. Cathepsin B downregulation reduced reactive oxygen species (ROS) accumulation and ERSID without affecting the induction of the unfolded protein response (UPR), but downregulation of PBA1 increased UPR and ERSID. Tonoplast rupture was not altered in the cathepsin B mutant and cathepsin B activation was independent of vacuolar processing enzyme (VPE). VPE activity was independent of cathepsin B. ERSID is regulated positively by cathepsin B and negatively by PBA1, revealing a complex picture behind caspase-3-like activity in plants. Cathepsin B may execute its function after tonoplast rupture and works in parallel with VPE.
Keywords: ER-stress-induced death (ERSID); endoplasmic reticulum (ER)-stress; plant protease; proteasome subunit PBA1; tunicamycin; unfolded protein response (UPR); vacuolar processing enzyme (VPE); vacuole.
© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
Figures


Comment in
-
Unravelling the mode of action of plant proteases.New Phytol. 2018 May;218(3):879-881. doi: 10.1111/nph.15156. New Phytol. 2018. PMID: 29658638 No abstract available.
References
-
- Cacas JL. 2010. Devil inside: does plant programmed cell death involve the endomembrane system? Plant, Cell & Environment 33: 1453–1473. - PubMed
-
- Costa MDL, Reis PAB, Valente MAS, Irsigler AST, Carvalho CM, Loureiro ME, Aragão FJL, Boston RS, Fietto LG, Fontes EPB. 2008. A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant‐specific asparagine‐rich proteins to promote cell death. Journal of Biological Chemistry 283: 20209–20219. - PubMed
-
- Craiu A, Gaczynska M, Akopian T, Gramm CF, Fenteany G, Goldberg AL, Rock KL. 1997. Lactacystin and clasto‐lactacystin beta‐lactone modify multiple proteasome beta‐subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. Journal of Biological Chemistry 272: 13437–13445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

