Separate and coincident expression of Hes1 and Hes5 in the developing mouse eye
- PMID: 28675662
- PMCID: PMC5739946
- DOI: 10.1002/dvdy.24542
Separate and coincident expression of Hes1 and Hes5 in the developing mouse eye
Abstract
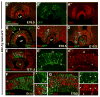
Background: Notch signaling is broadly required during embryogenesis, frequently activating the transcription of two basic helix-loop-helix transcription factors, Hes1 and Hes5. But, it remains unresolved when and where Hes1 and Hes5 act alone or together during development. Here, we analyzed a Hes5-green fluorescent protein (GFP) bacterial artificial chromosome (BAC) transgenic mouse, as a proxy for endogenous Hes5. We directly compared transgenic GFP expression with Hes1, and particular markers of embryonic lens and retina development.
Results: Hes5-GFP is dynamic within subsets of retinal and lens progenitor cells, and differentiating retinal ganglion neurons, in contrast to Hes1 found in all progenitor cells. In the adult retina, only Müller glia express Hes5-GFP. Finally, Hes5-GFP is up-regulated in Hes1 germline mutants, consistent with previous demonstration that Hes1 suppresses Hes5 transcription.
Conclusions: Hes5-GFP BAC transgenic mice are useful for identifying Hes5-expressing cells. Although Hes5-GFP and Hes1 are coexpressed in particular developmental contexts, we also noted cohorts of lens or retinal cells expressing just one factor. The dynamic Hes5-GFP expression pattern, coupled with its derepressed expression in Hes1 mutants, suggests that this transgene contains the relevant cis-regulatory elements that regulate endogenous Hes5 in the mouse lens and retina. Developmental Dynamics 247:212-221, 2018. © 2017 Wiley Periodicals, Inc.
Keywords: Hes1; Hes5; Notch signaling; lens development; retina, neurogenesis.
© 2017 Wiley Periodicals, Inc.
Figures




References
-
- Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–2476. - PubMed
-
- Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. - PubMed
-
- Bernardos RL, Lentz SI, Wolfe MS, Raymond PA. Notch-Delta signaling is required for spatial patterning and Muller glia differentiation in the zebrafish retina. Dev Biol. 2005;278:381–395. - PubMed
-
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

