Toward the identification of viral cap-methyltransferase inhibitors by fluorescence screening assay
- PMID: 28676301
- PMCID: PMC7113892
- DOI: 10.1016/j.antiviral.2017.06.021
Toward the identification of viral cap-methyltransferase inhibitors by fluorescence screening assay
Abstract
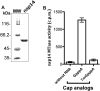
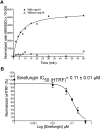
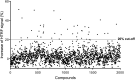
Two highly pathogenic human coronaviruses associated with severe respiratory syndromes emerged since the beginning of the century. The severe acute respiratory syndrome SARS-coronavirus (CoV) spread first in southern China in 2003 with about 8000 infected cases in few months. Then in 2012, the Middle East respiratory syndrome (MERS-CoV) emerged from the Arabian Peninsula giving a still on-going epidemic associated to a high fatality rate. CoVs are thus considered a major health threat. This is especially true as no vaccine nor specific therapeutic are available against either SARS- or MERS-CoV. Therefore, new drugs need to be identified in order to develop antiviral treatments limiting CoV replication. In this study, we focus on the nsp14 protein, which plays a key role in virus replication as it methylates the RNA cap structure at the N7 position of the guanine. We developed a high-throughput N7-MTase assay based on Homogenous Time Resolved Fluorescence (HTRF®) and screened chemical libraries (2000 compounds) on the SARS-CoV nsp14. 20 compounds inhibiting the SARS-CoV nsp14 were further evaluated by IC50 determination and their specificity was assessed toward flavivirus- and human cap N7-MTases. Our results reveal three classes of compounds: 1) molecules inhibiting several MTases as well as the dengue virus polymerase activity unspecifically, 2) pan MTases inhibitors targeting both viral and cellular MTases, and 3) inhibitors targeting one viral MTase more specifically showing however activity against the human cap N7-MTase. These compounds provide a first basis towards the development of more specific inhibitors of viral methyltransferases.
Keywords: Antiviral; Coronavirus; Flavivirus; HTRF; Inhibitor; Methyltransferase.
Copyright © 2017. Published by Elsevier B.V.
Figures
References
-
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3 ’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. PNAS. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109/-/DCSupplemental. www.pnas.org/cgi/doi/10.1073/pnas.1201130109 - DOI - PMC - PubMed
-
- Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lécine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. - DOI - PMC - PubMed
-
- Case J.B., Ashbrook A.W., Dermody T.S., Denison M.R. Mutagenesis of S-Adenosyl-L-Methionine-Binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90:7248–7256. doi: 10.1128/JVI.00542-16. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

