Kinetic and structural analyses reveal residues in phosphoinositide 3-kinase α that are critical for catalysis and substrate recognition
- PMID: 28676499
- PMCID: PMC5566514
- DOI: 10.1074/jbc.M116.772426
Kinetic and structural analyses reveal residues in phosphoinositide 3-kinase α that are critical for catalysis and substrate recognition
Abstract
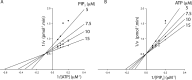
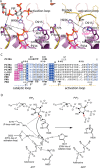
Phosphoinositide 3-kinases (PI3Ks) are ubiquitous lipid kinases that activate signaling cascades controlling cell survival, proliferation, protein synthesis, and vesicle trafficking. PI3Ks have dual kinase specificity: a lipid kinase activity that phosphorylates the 3'-hydroxyl of phosphoinositides and a protein-kinase activity that includes autophosphorylation. Despite the wealth of biochemical and structural information on PI3Kα, little is known about the identity and roles of individual active-site residues in catalysis. To close this gap, we explored the roles of residues of the catalytic domain and the regulatory subunit of human PI3Kα in lipid and protein phosphorylation. Using site-directed mutagenesis, kinetic assays, and quantitative mass spectrometry, we precisely mapped key residues involved in substrate recognition and catalysis by PI3Kα. Our results revealed that Lys-776, located in the P-loop of PI3Kα, is essential for the recognition of lipid and ATP substrates and also plays an important role in PI3Kα autophosphorylation. Replacement of the residues His-936 and His-917 in the activation and catalytic loops, respectively, with alanine dramatically changed PI3Kα kinetics. Although H936A inactivated the lipid kinase activity without affecting autophosphorylation, H917A abolished both the lipid and protein kinase activities of PI3Kα. On the basis of these kinetic and structural analyses, we propose possible mechanistic roles of these critical residues in PI3Kα catalysis.
Keywords: ATP; lipid kinase; p110a; p110ap85a; phosphatidylinositide 3-kinase (PI 3-kinase); phosphoinositide; protein kinase; serine/threonine protein kinase.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Fruman D. A., Meyers R. E., and Cantley L. C. (1998) Phosphoinositide kinases. Annu. Rev. Biochem. 67, 481–507 - PubMed
-
- Chang H. W., Aoki M., Fruman D., Auger K. R., Bellacosa A., Tsichlis P. N., Cantley L. C., Roberts T. M., and Vogt P. K. (1997) Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science 276, 1848–1850 - PubMed
-
- Cho H., Mu J., Kim J. K., Thorvaldsen J. L., Chu Q., Crenshaw E. B. 3rd, Kaestner K. H., Bartolomei M. S., Shulman G. I., and Birnbaum M. J. (2001) Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 292, 1728–1731 - PubMed
-
- Maira S. M., Finan P., and Garcia-Echeverria C. (2010) From the bench to the bed side: PI3K pathway inhibitors in clinical development. Curr. Top. Microbiol. Immunol. 347, 209–239 - PubMed
-
- Koorella C., Nair J. R., Murray M. E., Carlson L. M., Watkins S. K., and Lee K. P. (2014) Novel regulation of CD80/CD86-induced phosphatidylinositol 3-kinase signaling by NOTCH1 protein in interleukin-6 and indoleamine 2,3-dioxygenase production by dendritic cells. J. Biol. Chem. 289, 7747–7762 - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

