Functional consequences of the first reported mutations of the proto-oncogene PTTG1IP/PBF
- PMID: 28676500
- PMCID: PMC5551380
- DOI: 10.1530/ERC-16-0340
Functional consequences of the first reported mutations of the proto-oncogene PTTG1IP/PBF
Abstract
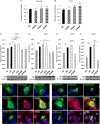
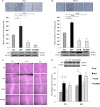
Pituitary tumor-transforming gene 1-binding factor (PTTG1IP; PBF) is a multifunctional glycoprotein, which is overexpressed in a wide range of tumours, and significantly associated with poorer oncological outcomes, such as early tumour recurrence, distant metastasis, extramural vascular invasion and decreased disease-specific survival. PBF transforms NIH 3T3 fibroblasts and induces tumours in nude mice, while mice harbouring transgenic thyroidal PBF expression show hyperplasia and macrofollicular lesions. Our assumption that PBF becomes an oncogene purely through increased expression has been challenged by the recent report of mutations in PBF within the Catalogue of Somatic Mutations in Cancer (COSMIC) database. We therefore sought to determine whether the first 10 PBF missense substitutions in human cancer might be oncogenic. Anisomycin half-life studies revealed that most mutations were associated with reduced protein stability compared to wild-type (WT) PBF. Proliferation assays narrowed our interest to two mutational events which significantly altered cell turnover: C51R and R140W. C51R was mainly confined to the endoplasmic reticulum while R140W was apparent in the Golgi apparatus. Both C51R and R140W lost the capacity to induce cellular migration and significantly reduced cell invasion. Colony formation and soft agar assays demonstrated that, in contrast to WT PBF, both mutants were unable to elicit significant colony formation or anchorage-independent growth. However, C51R and R140W retained the ability to repress radioiodide uptake, a functional hallmark of PBF. Our data reveal new insight into PBF function and confirm that, rather than being oncogenic, mutations in PBF are likely to be passenger effects, with overexpression of PBF the more important aetiological event in human cancer.
Keywords: COSMIC; PTTG1IP; TCGA; mutation; proliferation.
© 2017 The authors.
Figures




References
-
- Boelaert K, McCabe CJ, Tannahill LA, Gittoes NJ, Holder RL, Watkinson JC, Bradwell AR, Sheppard MC, Franklyn JA. 2003. Pituitary tumor transforming gene and fibroblast growth factor-2 expression: potential prognostic indicators in differentiated thyroid cancer. Journal of Clinical Endocrinology and Metabolism 88 2341–2347. (10.1210/jc.2002-021113) - DOI - PubMed
-
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2 401–404. (10.1158/2159-8290.CD-12-0095) - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

