Sorghum Dw2 Encodes a Protein Kinase Regulator of Stem Internode Length
- PMID: 28676627
- PMCID: PMC5496852
- DOI: 10.1038/s41598-017-04609-5
Sorghum Dw2 Encodes a Protein Kinase Regulator of Stem Internode Length
Abstract
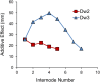
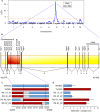
Sorghum is an important C4 grass crop grown for grain, forage, sugar, and bioenergy production. While tall, late flowering landraces are commonly grown in Africa, short early flowering varieties were selected in US grain sorghum breeding programs to reduce lodging and to facilitate machine harvesting. Four loci have been identified that affect stem length (Dw1-Dw4). Subsequent research showed that Dw3 encodes an ABCB1 auxin transporter and Dw1 encodes a highly conserved protein involved in the regulation of cell proliferation. In this study, Dw2 was identified by fine-mapping and further confirmed by sequencing the Dw2 alleles in Dwarf Yellow Milo and Double Dwarf Yellow Milo, the progenitor genotypes where the recessive allele of dw2 originated. The Dw2 locus was determined to correspond to Sobic.006G067700, a gene that encodes a protein kinase that is homologous to KIPK, a member of the AGCVIII subgroup of the AGC protein kinase family in Arabidopsis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Quinby, J. Sorghum Improvement and the Genetics of Growth. (Texas A&M University Press, 1974).
-
- Murray SC, et al. Genetic improvement of sorghum as a biofuel feedstock: I. QTL for stem sugar and grain nonstructural carbohydrates. Crop Sci. 2008;48:2165–2179. doi: 10.2135/cropsci2008.01.0016. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

