Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans
- PMID: 28677677
- PMCID: PMC5504273
- DOI: 10.1038/ncomms15955
Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans
Abstract
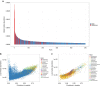
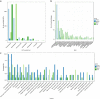
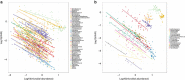
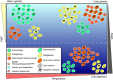
Marine viruses are key drivers of host diversity, population dynamics and biogeochemical cycling and contribute to the daily flux of billions of tons of organic matter. Despite recent advancements in metagenomics, much of their biodiversity remains uncharacterized. Here we report a data set of 27,346 marine virome contigs that includes 44 complete genomes. These outnumber all currently known phage genomes in marine habitats and include members of previously uncharacterized lineages. We designed a new method for host prediction based on co-occurrence associations that reveals these viruses infect dominant members of the marine microbiome such as Prochlorococcus and Pelagibacter. A negative association between host abundance and the virus-to-host ratio supports the recently proposed Piggyback-the-Winner model of reduced phage lysis at higher host densities. An analysis of the abundance patterns of viruses throughout the oceans revealed how marine viral communities adapt to various seasonal, temperature and photic regimes according to targeted hosts and the diversity of auxiliary metabolic genes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Suttle C. A. Viruses in the sea. Nature 437, 356–361 (2005). - PubMed
-
- Breitbart M. Marine viruses: truth or dare. Mar. Sci. 4, 425–448 (2012). - PubMed
-
- Brussaard C. P. D. et al.. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2, 575–578 (2008). - PubMed
-
- Suttle C. A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007). - PubMed
-
- Wilhelm W. & Suttle C. A. Viruses and nutrient cycles in the sea. Bioscience 49, 781–788 (1999).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

