HIV Env conserved element DNA vaccine alters immunodominance in macaques
- PMID: 28678607
- PMCID: PMC5718827
- DOI: 10.1080/21645515.2017.1339852
HIV Env conserved element DNA vaccine alters immunodominance in macaques
Abstract
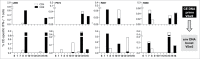
Sequence diversity and immunodominance are major obstacles in the design of an effective vaccine against HIV. HIV Env is a highly-glycosylated protein composed of 'conserved' and 'variable' regions. The latter contains immunodominant epitopes that are frequently targeted by the immune system resulting in the generation of immune escape variants. This work describes 12 regions in HIV Env that are highly conserved throughout the known HIV M Group sequences (Env CE), and are poorly immunogenic in macaques vaccinated with full-length Env expressing DNA vaccines. Two versions of plasmids encoding the 12 Env CE were generated, differing by 0-5 AA per CE to maximize the inclusion of commonly detected variants. In contrast to the full-length env DNA vaccine, vaccination of macaques with a combination of these 2 Env CE DNA induced robust, durable cellular immune responses with a significant fraction of CD8+ T cells with cytotoxic phenotype (Granzyme B+ and CD107a+). Although inefficient in generating primary responses to the CE, boosting of the Env CE DNA primed macaques with the intact env DNA vaccine potently augmented pre-existing immunity, increasing magnitude, breadth and cytotoxicity of the cellular responses. Fine mapping showed that 7 of the 12 CE elicited T cell responses. Env CE DNA also induced humoral responses able to recognize the full-length Env. Env CE plasmids are therefore capable of inducing durable responses to highly conserved regions of Env that are frequently absent after Env vaccination or immunologically subdominant. These modified antigens are candidates for use as prophylactic and therapeutic HIV vaccines.
Keywords: DNA vaccine; Env; HIV; conserved epitopes; cytotoxic T cells; immunization; prime-boost; rhesus macaque; subdominant epitopes; variable epitopes.
Figures






References
-
- Carlson JM, Brumme CJ, Martin E, Listgarten J, Brockman MA, Le AQ, Chui CK, Cotton LA, Knapp DJ, Riddler SA, et al.. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol. 2012;86:13202-16. doi: 10.1128/JVI.01998-12. PMID:23055555 - DOI - PMC - PubMed
-
- Keane NM, Roberts SG, Almeida CA, Krishnan T, Chopra A, Demaine E, Laird R, Tschochner M, Carlson JM, Mallal S, et al.. High-avidity, high-IFNgamma-producing CD8 T-cell responses following immune selection during HIV-1 infection. Immunol Cell Biol. 2012;90:224-34. doi: 10.1038/icb.2011.34. PMID:21577229 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
