Crystal structures of agonist-bound human cannabinoid receptor CB1
- PMID: 28678776
- PMCID: PMC5793864
- DOI: 10.1038/nature23272
Crystal structures of agonist-bound human cannabinoid receptor CB1
Retraction in
-
Retraction Note: Crystal structures of agonist-bound human cannabinoid receptor CB1.Nature. 2025 Sep;645(8080):544. doi: 10.1038/s41586-025-09538-2. Nature. 2025. PMID: 40866714 No abstract available.
Abstract
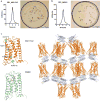
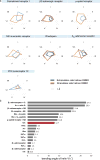
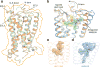
The cannabinoid receptor 1 (CB1) is the principal target of the psychoactive constituent of marijuana, the partial agonist Δ9-tetrahydrocannabinol (Δ9-THC). Here we report two agonist-bound crystal structures of human CB1 in complex with a tetrahydrocannabinol (AM11542) and a hexahydrocannabinol (AM841) at 2.80 Å and 2.95 Å resolution, respectively. The two CB1-agonist complexes reveal important conformational changes in the overall structure, relative to the antagonist-bound state, including a 53% reduction in the volume of the ligand-binding pocket and an increase in the surface area of the G-protein-binding region. In addition, a 'twin toggle switch' of Phe2003.36 and Trp3566.48 (superscripts denote Ballesteros-Weinstein numbering) is experimentally observed and appears to be essential for receptor activation. The structures reveal important insights into the activation mechanism of CB1 and provide a molecular basis for predicting the binding modes of Δ9-THC, and endogenous and synthetic cannabinoids. The plasticity of the binding pocket of CB1 seems to be a common feature among certain class A G-protein-coupled receptors. These findings should inspire the design of chemically diverse ligands with distinct pharmacological properties.
Conflict of interest statement
The authors declare competing financial interests: details are available in the online version of the paper.
Figures







References
-
- Mechoulam R, Hanuš LO, Pertwee R, Howlett AC. Early phytocannabinoid chemistry to endocannabinoids and beyond. Nat. Rev. Neurosci. 2014;15:757–764. - PubMed
-
- Ballesteros JA, Weinstein H. In: Methods in Neurosciences. Sealfon Stuart C, editor. Vol. 25. Academic; 1995. pp. 366–428.
-
- Lemberger L. Potential therapeutic usefulness of marijuana. Annu. Rev. Pharmacol. Toxicol. 1980;20:151–172. - PubMed
-
- Li H-L. An archaeological and historical account of cannabis in China. Econ. Bot. 1973;28:437–448.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

