Nutrient sensing modulates malaria parasite virulence
- PMID: 28678779
- PMCID: PMC5511512
- DOI: 10.1038/nature23009
Nutrient sensing modulates malaria parasite virulence
Abstract
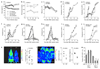
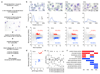
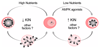
The lifestyle of intracellular pathogens, such as malaria parasites, is intimately connected to that of their host, primarily for nutrient supply. Nutrients act not only as primary sources of energy but also as regulators of gene expression, metabolism and growth, through various signalling networks that enable cells to sense and adapt to varying environmental conditions. Canonical nutrient-sensing pathways are presumed to be absent from the causative agent of malaria, Plasmodium, thus raising the question of whether these parasites can sense and cope with fluctuations in host nutrient levels. Here we show that Plasmodium blood-stage parasites actively respond to host dietary calorie alterations through rearrangement of their transcriptome accompanied by substantial adjustment of their multiplication rate. A kinome analysis combined with chemical and genetic approaches identified KIN as a critical regulator that mediates sensing of nutrients and controls a transcriptional response to the host nutritional status. KIN shares homology with SNF1/AMPKα, and yeast complementation studies suggest that it is part of a functionally conserved cellular energy-sensing pathway. Overall, these findings reveal a key parasite nutrient-sensing mechanism that is critical for modulating parasite replication and virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




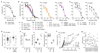


Comment in
-
Malaria Parasites in a Hungry Host: Kinases and Host Caloric Restriction Brought Together.Trends Parasitol. 2017 Nov;33(11):831-832. doi: 10.1016/j.pt.2017.08.007. Epub 2017 Sep 8. Trends Parasitol. 2017. PMID: 28923378
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

