Dynamical models of mutated chronic myelogenous leukemia cells for a post-imatinib treatment scenario: Response to dasatinib or nilotinib therapy
- PMID: 28678800
- PMCID: PMC5497988
- DOI: 10.1371/journal.pone.0179700
Dynamical models of mutated chronic myelogenous leukemia cells for a post-imatinib treatment scenario: Response to dasatinib or nilotinib therapy
Abstract
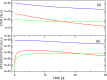
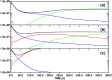
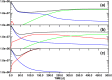
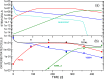
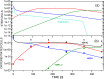
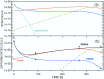
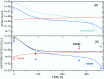
Targeted inhibition of the oncogenic BCR-ABL1 fusion protein using the ABL1 tyrosine kinase inhibitor imatinib has become standard therapy for chronic myelogenous leukemia (CML), with most patients reaching total and durable remission. However, a significant fraction of patients develop resistance, commonly due to mutated ABL1 kinase domains. This motivated development of second-generation drugs with broadened or altered protein kinase selectivity profiles, including dasatinib and nilotinib. Imatinib-resistant patients undergoing treatment with second-line drugs typically develop resistance to them, but dynamic and clonal properties of this response differ. Shared, however, is the observation of clonal competition, reflected in patterns of successive dominance of individual clones. We present three deterministic mathematical models to study the origins of clinically observed dynamics. Each model is a system of coupled first-order differential equations, considering populations of three mutated active stem cell strains and three associated pools of differentiated cells; two models allow for activation of quiescent stem cells. Each approach is distinguished by the way proliferation rates of the primary stem cell reservoir are modulated. Previous studies have concentrated on simulating the response of wild-type leukemic cells to imatinib administration; our focus is on modelling the time dependence of imatinib-resistant clones upon subsequent exposure to dasatinib or nilotinib. Performance of the three computational schemes to reproduce selected CML patient profiles is assessed. While some simple cases can be approximated by a basic design that does not invoke quiescence, others are more complex and require involvement of non-cycling stem cells for reproduction. We implement a new feedback mechanism for regulation of coupling between cycling and non-cycling stem cell reservoirs that depends on total cell populations. A bifurcation landscape analysis is also performed for solutions to the basic ansatz. Computational models reproducing patient data illustrate potential dynamic mechanisms that may guide optimization of therapy of drug resistant CML.
Conflict of interest statement
Figures







Similar articles
-
Frequency of ABL gene mutations in chronic myeloid leukemia patients resistant to imatinib and results of treatment switch to second-generation tyrosine kinase inhibitors.Med Clin (Barc). 2013 Aug 4;141(3):95-9. doi: 10.1016/j.medcli.2012.10.028. Epub 2013 Feb 22. Med Clin (Barc). 2013. PMID: 23433665
-
Sequential inhibitor therapy in CML: in vitro simulation elucidates the pattern of resistance mutations after second- and third-line treatment.Clin Cancer Res. 2013 Jun 1;19(11):2962-72. doi: 10.1158/1078-0432.CCR-13-0052. Epub 2013 Apr 2. Clin Cancer Res. 2013. PMID: 23549879
-
Improving frontline treatment for chronic myeloid leukemia: emerging evidence for use of nilotinib and dasatinib.Clin Adv Hematol Oncol. 2011 Oct;9(10):734-45. Clin Adv Hematol Oncol. 2011. PMID: 22252576 Review.
-
Dynamics of chronic myeloid leukemia response to dasatinib, nilotinib, and high-dose imatinib.Haematologica. 2014 Nov;99(11):1701-9. doi: 10.3324/haematol.2013.085977. Epub 2014 Sep 12. Haematologica. 2014. PMID: 25216683 Free PMC article.
-
Characterization of cancer stem cells in chronic myeloid leukaemia.Biochem Soc Trans. 2007 Nov;35(Pt 5):1347-51. doi: 10.1042/BST0351347. Biochem Soc Trans. 2007. PMID: 17956348 Review.
Cited by
-
Computational modeling reveals key factors driving treatment-free remission in chronic myeloid leukemia patients.NPJ Syst Biol Appl. 2024 Apr 27;10(1):45. doi: 10.1038/s41540-024-00370-4. NPJ Syst Biol Appl. 2024. PMID: 38678088 Free PMC article.
-
The impact of introducing tyrosine kinase inhibitors on chronic myeloid leukemia survival: a population-based study.BMC Cancer. 2018 Nov 6;18(1):1069. doi: 10.1186/s12885-018-4984-3. BMC Cancer. 2018. PMID: 30400842 Free PMC article.
-
Predictive nonlinear modeling of malignant myelopoiesis and tyrosine kinase inhibitor therapy.Elife. 2023 Apr 28;12:e84149. doi: 10.7554/eLife.84149. Elife. 2023. PMID: 37115622 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

