KSHV encoded ORF59 modulates histone arginine methylation of the viral genome to promote viral reactivation
- PMID: 28678843
- PMCID: PMC5513536
- DOI: 10.1371/journal.ppat.1006482
KSHV encoded ORF59 modulates histone arginine methylation of the viral genome to promote viral reactivation
Abstract
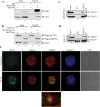
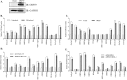
Kaposi's sarcoma associated herpesvirus (KSHV) persists in a highly-ordered chromatin structure inside latently infected cells with the majority of the viral genome having repressive marks. However, upon reactivation the viral chromatin landscape changes into 'open' chromatin through the involvement of lysine demethylases and methyltransferases. Besides methylation of lysine residues of histone H3, arginine methylation of histone H4 plays an important role in controlling the compactness of the chromatin. Symmetric methylation of histone H4 at arginine 3 (H4R3me2s) negatively affects the methylation of histone H3 at lysine 4 (H3K4me3), an active epigenetic mark deposited on the viral chromatin during reactivation. We identified a novel binding partner to KSHV viral DNA processivity factor, ORF59-a protein arginine methyl transferase 5 (PRMT5). PRMT5 is an arginine methyltransferase that dimethylates arginine 3 (R3) of histone H4 in a symmetric manner, one hallmark of condensed chromatin. Our ChIP-seq data of symmetrically methylated H4 arginine 3 showed a significant decrease in H4R3me2s on the viral genome of reactivated cells as compared to the latent cells. Reduction in arginine methylation correlated with the binding of ORF59 on the viral chromatin and disruption of PRMT5 from its adapter protein, COPR5 (cooperator of PRMT5). Binding of PRMT5 through COPR5 is important for symmetric methylation of H4R3 and the expression of ORF59 competitively reduces the association of PRMT5 with COPR5, leading to a reduction in PRMT5 mediated arginine methylation. This ultimately resulted in a reduced level of symmetrically methylated H4R3 and increased levels of H3K4me3 marks, contributing to the formation of an open chromatin for transcription and DNA replication. Depletion of PRMT5 levels led to a decrease in symmetric methylation and increase in viral gene transcription confirming the role of PRMT5 in viral reactivation. In conclusion, ORF59 modulates histone-modifying enzymes to alter the chromatin structure during lytic reactivation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
KSHV ORF59 and PAN RNA Recruit Histone Demethylases to the Viral Chromatin during Lytic Reactivation.Viruses. 2020 Apr 9;12(4):420. doi: 10.3390/v12040420. Viruses. 2020. PMID: 32283586 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Processivity Factor, ORF59, Binds to Canonical and Linker Histones, and Its Carboxy Terminus Is Dispensable for Viral DNA Synthesis.J Virol. 2021 Feb 24;95(6):e02169-20. doi: 10.1128/JVI.02169-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361421 Free PMC article.
-
OGG1 regulates the level of symmetric dimethylation of histone H4 arginine-3 by interacting with PRMT5.Mol Cell Probes. 2018 Apr;38:19-24. doi: 10.1016/j.mcp.2018.01.002. Epub 2018 Feb 2. Mol Cell Probes. 2018. PMID: 29409673
-
An atlas of chromatin landscape in KSHV-infected cells during de novo infection and reactivation.Virology. 2024 Sep;597:110146. doi: 10.1016/j.virol.2024.110146. Epub 2024 Jun 19. Virology. 2024. PMID: 38909515 Review.
-
Molecular biology of KSHV lytic reactivation.Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review.
Cited by
-
Protein Arginine Methyltransferase 5 Functions via Interacting Proteins.Front Cell Dev Biol. 2021 Aug 27;9:725301. doi: 10.3389/fcell.2021.725301. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513846 Free PMC article.
-
Transcriptional and post-transcriptional regulation of viral gene expression in the gamma-herpesvirus Kaposi's sarcoma-associated herpesvirus.Curr Clin Microbiol Rep. 2018 Dec;5(4):219-228. doi: 10.1007/s40588-018-0102-1. Epub 2018 Aug 3. Curr Clin Microbiol Rep. 2018. PMID: 30854283 Free PMC article.
-
Cellular consequences of arginine methylation.Cell Mol Life Sci. 2019 Aug;76(15):2933-2956. doi: 10.1007/s00018-019-03140-2. Epub 2019 May 17. Cell Mol Life Sci. 2019. PMID: 31101937 Free PMC article. Review.
-
Analysis of Metabolomic Reprogramming Induced by Infection with Kaposi's Sarcoma-Associated Herpesvirus Using Untargeted Metabolomic Profiling.Int J Mol Sci. 2025 Mar 28;26(7):3109. doi: 10.3390/ijms26073109. Int J Mol Sci. 2025. PMID: 40243754 Free PMC article.
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
References
-
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–91. doi: 10.1056/NEJM199505043321802 . - DOI - PubMed
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266(5192):1865–9. Epub 1994/12/16. . - PubMed
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86(4):1276–80. . - PubMed
-
- Uldrick TS, Wang V, O'Mahony D, Aleman K, Wyvill KM, Marshall V, et al. An Interleukin-6-Related Systemic Inflammatory Syndrome in Patients Co-Infected with Kaposi Sarcoma-Associated Herpesvirus and HIV but without Multicentric Castleman Disease. Clin Infect Dis. 2010;51(3):350–8. Epub 2010/06/30. doi: 10.1086/654798 . - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

