Q&A: using Patch-seq to profile single cells
- PMID: 28679385
- PMCID: PMC5499043
- DOI: 10.1186/s12915-017-0396-0
Q&A: using Patch-seq to profile single cells
Abstract
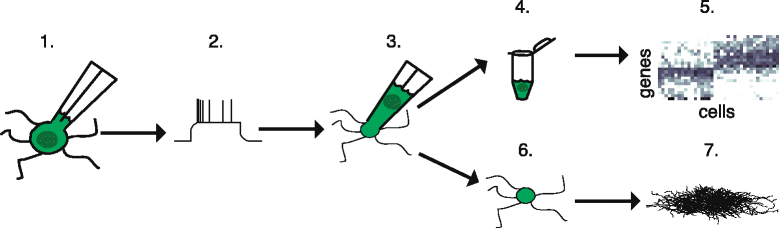
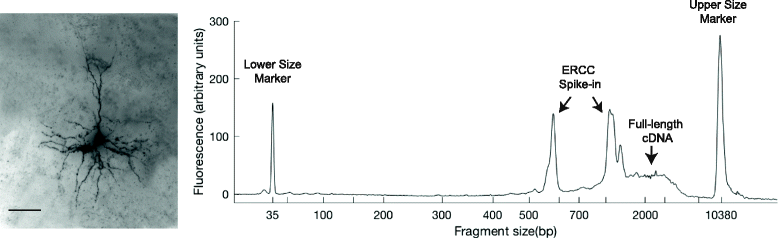
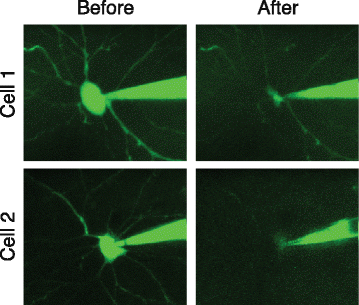
Individual neurons vary widely in terms of their gene expression, morphology, and electrophysiological properties. While many techniques exist to study single-cell variability along one or two of these dimensions, very few techniques can assess all three features for a single cell. We recently developed Patch-seq, which combines whole-cell patch clamp recording with single-cell RNA-sequencing and immunohistochemistry to comprehensively profile the transcriptomic, morphologic, and physiologic features of individual neurons. Patch-seq can be broadly applied to characterize cell types in complex tissues such as the nervous system, and to study the transcriptional signatures underlying the multidimensional phenotypes of single cells.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

