Data normalization considerations for digital tumor dissection
- PMID: 28679399
- PMCID: PMC5498978
- DOI: 10.1186/s13059-017-1257-4
Data normalization considerations for digital tumor dissection
Abstract
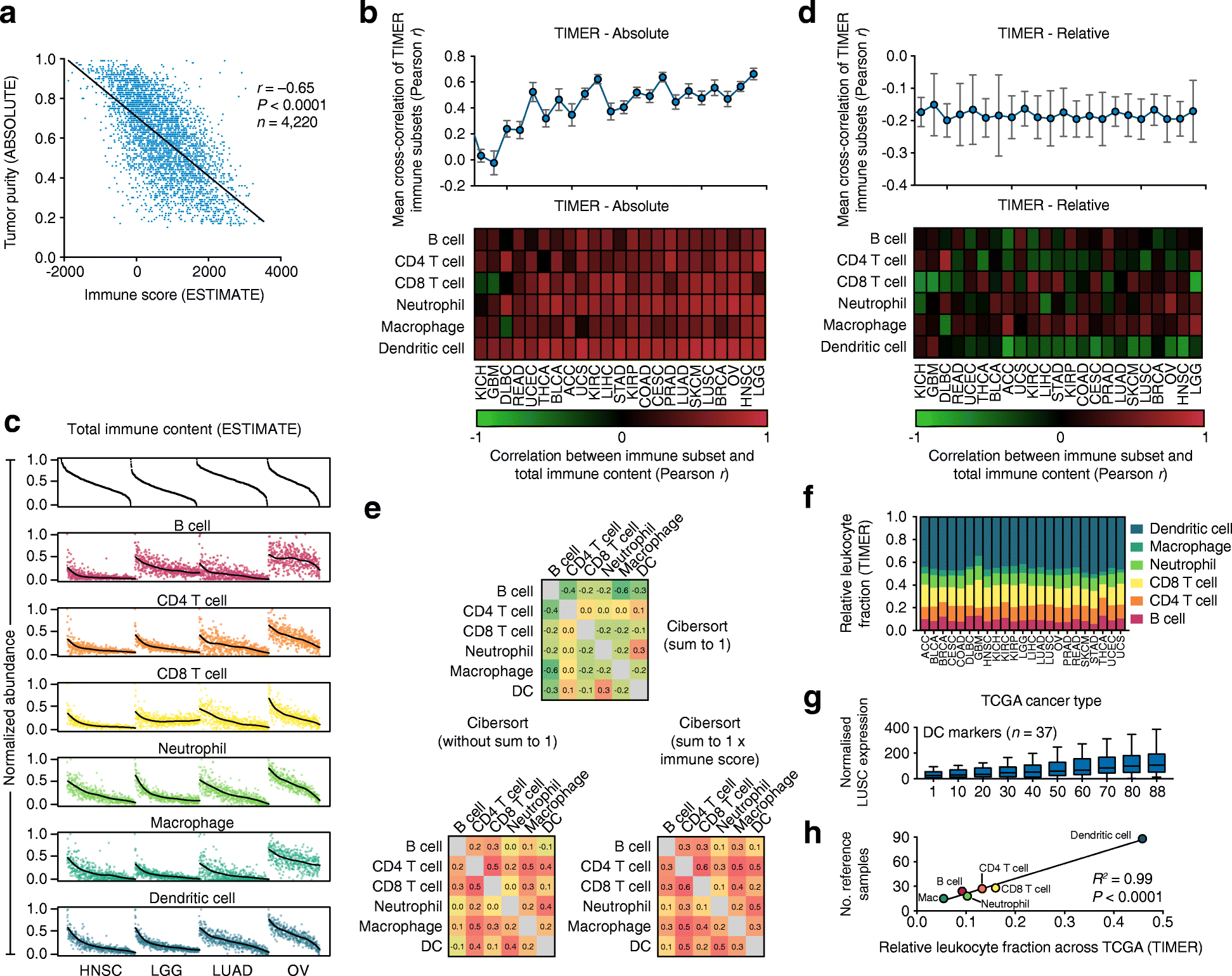
In a recently published article in Genome Biology, Li and colleagues introduced TIMER, a gene expression deconvolution approach for studying tumor-infiltrating leukocytes (TILs) in 23 cancer types profiled by The Cancer Genome Atlas. Methods to characterize TIL biology are increasingly important, and the authors offer several arguments in favor of their strategy. Several of these claims warrant further discussion and highlight the critical importance of data normalization in gene expression deconvolution applications.Please see related Li et al correspondence: www.dx.doi.org/10.1186/s13059-017-1256-5 and Zheng correspondence: www.dx.doi.org/10.1186/s13059-017-1258-3.
Figures

Comment in
-
Revisit linear regression-based deconvolution methods for tumor gene expression data.Genome Biol. 2017 Jul 5;18(1):127. doi: 10.1186/s13059-017-1256-5. Genome Biol. 2017. PMID: 28679386 Free PMC article. No abstract available.
Comment on
-
Comprehensive analyses of tumor immunity: implications for cancer immunotherapy.Genome Biol. 2016 Aug 22;17(1):174. doi: 10.1186/s13059-016-1028-7. Genome Biol. 2016. PMID: 27549193 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

