Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis
- PMID: 28679656
- PMCID: PMC5706545
- DOI: 10.1126/scitranslmed.aal4651
Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis
Abstract
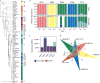
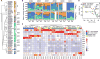
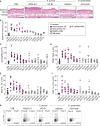
The heterogeneous course, severity, and treatment responses among patients with atopic dermatitis (AD; eczema) highlight the complexity of this multifactorial disease. Prior studies have used traditional typing methods on cultivated isolates or sequenced a bacterial marker gene to study the skin microbial communities of AD patients. Shotgun metagenomic sequence analysis provides much greater resolution, elucidating multiple levels of microbial community assembly ranging from kingdom to species and strain-level diversification. We analyzed microbial temporal dynamics from a cohort of pediatric AD patients sampled throughout the disease course. Species-level investigation of AD flares showed greater Staphylococcus aureus predominance in patients with more severe disease and Staphylococcus epidermidis predominance in patients with less severe disease. At the strain level, metagenomic sequencing analyses demonstrated clonal S. aureus strains in more severe patients and heterogeneous S. epidermidis strain communities in all patients. To investigate strain-level biological effects of S. aureus, we topically colonized mice with human strains isolated from AD patients and controls. This cutaneous colonization model demonstrated S. aureus strain-specific differences in eliciting skin inflammation and immune signatures characteristic of AD patients. Specifically, S. aureus isolates from AD patients with more severe flares induced epidermal thickening and expansion of cutaneous T helper 2 (TH2) and TH17 cells. Integrating high-resolution sequencing, culturing, and animal models demonstrated how functional differences of staphylococcal strains may contribute to the complexity of AD disease.
Copyright © 2017 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


References
-
- Eyerich K, Eyerich S, Biedermann T. The Multi-Modal Immune Pathogenesis of Atopic Eczema. Trends in immunology. 2015;36:788–801. - PubMed
-
- E. A. Genetics, C. Lifecourse Epidemiology Eczema, C. Australian Asthma Genetics, A. Australian Asthma Genetics Consortium. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nature genetics. 2015;47:1449–1456. - PMC - PubMed
-
- Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O’Regan GM, Watson RM, Cecil JE, Bale SJ, Compton JG, DiGiovanna JJ, Fleckman P, Lewis-Jones S, Arseculeratne G, Sergeant A, Munro CS, El Houate B, McElreavey K, Halkjaer LB, Bisgaard H, Mukhopadhyay S, McLean WH. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nature genetics. 2006;38:441–446. - PubMed
-
- Huang JT, Abrams M, Tlougan B, Rademaker A, Paller AS. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–814. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

