Structural properties of genotype-phenotype maps
- PMID: 28679667
- PMCID: PMC5550974
- DOI: 10.1098/rsif.2017.0275
Structural properties of genotype-phenotype maps
Abstract
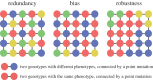
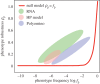
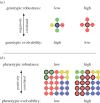
The map between genotype and phenotype is fundamental to biology. Biological information is stored and passed on in the form of genotypes, and expressed in the form of phenotypes. A growing body of literature has examined a wide range of genotype-phenotype (GP) maps and has established a number of properties that appear to be shared by many GP maps. These properties are 'structural' in the sense that they are properties of the distribution of phenotypes across the point-mutation network of genotypes. They include: a redundancy of genotypes, meaning that many genotypes map to the same phenotypes, a highly non-uniform distribution of the number of genotypes per phenotype, a high robustness of phenotypes and the ability to reach a large number of new phenotypes within a small number of mutational steps. A further important property is that the robustness and evolvability of phenotypes are positively correlated. In this review, I give an overview of the study of GP maps with particular emphasis on these structural properties, and discuss a model that attempts to explain why these properties arise, as well as some of the fundamental ways in which the structure of GP maps can affect evolutionary outcomes.
Keywords: RNA secondary structure; evolvability; genotype; neutral evolution; phenotype; robustness.
© 2017 The Author(s).
Conflict of interest statement
I declare I have no competing interests.
Figures

References
-
- Mitchell M. 1996. An introduction to genetic algorithms. Cambridge, MA: MIT Press.
-
- Mendel G. 1866. Verhandlungen des naturforschenden Vereines in Brunn 4, 3–47.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
