An unusual genomic variant of pancreatic ductal adenocarcinoma with an indolent clinical course
- PMID: 28679692
- PMCID: PMC5495033
- DOI: 10.1101/mcs.a001701
An unusual genomic variant of pancreatic ductal adenocarcinoma with an indolent clinical course
Abstract
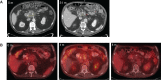
We describe an 85-yr-old male of Ashkenazi Jewish descent with biopsy-proven locally advanced pancreatic ductal adenocarcinoma (PDA). The patient underwent a modified course of gemcitabine and stereotactic body radiation therapy and survived for 42 mo with a stable pancreatic head mass and no evidence of metastatic disease before death due to complications from a stroke. Whole-exome sequencing of his tumor revealed a simple genome landscape with no evidence of mutations, copy-number changes, or structural alterations in genes most commonly associated with PDA (i.e., KRAS, CDKN2A, TP53, or SMAD4). An analysis of his germline DNA revealed no pathogenic variants of significance. Whole-exome and whole-genome sequencing identified a somatic mutation of RNF213 and an inversion/deletion of CTNNA2 as the genetic basis of his PDA. Although PDA is classically characterized by a predictable set of mutations, these data suggest that alternate genetic paths to PDA may exist, which can be associated with a more indolent clinical course.
Keywords: neoplasm of the pancreas; pancreatic adenocarcinoma.
© 2017 Kohutek et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, et al. 2016. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531: 47–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
