AIM2-Like Receptors Positively and Negatively Regulate the Interferon Response Induced by Cytosolic DNA
- PMID: 28679751
- PMCID: PMC5573678
- DOI: 10.1128/mBio.00944-17
AIM2-Like Receptors Positively and Negatively Regulate the Interferon Response Induced by Cytosolic DNA
Abstract
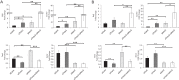
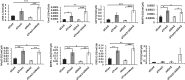
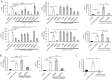
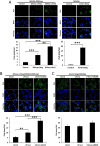
Cytosolic DNAs derived from retrotransposons serve as pathogen-associated molecular patterns for pattern recognition receptors (PRRs) that stimulate the induction of interferons (IFNs) and other cytokines, leading to autoimmune disease. Cyclic GMP-AMP synthase is one PRR that senses retrotransposon DNA, activating type I IFN responses through the stimulator of IFN genes (STING). Absent in melanoma 2 (AIM2)-like receptors (ALRs) have also been implicated in these pathways. Here we show that the mouse ALR IFI205 senses cytosolic retrotransposon DNA independently of cyclic GMP-AMP production. AIM2 antagonizes IFI205-mediated IFN induction activity by sequestering it from STING. We also found that the complement of genes located in the ALR locus in C57BL/6 and AIM2 knockout mice are different and unique, which has implications for interpretation of the sensing of pathogens in different mouse strains. Our data suggest that members of the ALR family are critical to the host IFN response to endogenous DNA.IMPORTANCE Autoimmune diseases like Aicardi-Goutières syndrome and lupus erythematosus arise when cells of the immune system become activated and attack host cells and tissues. We found that DNA generated by endogenous retroviruses and retroelements in inbred mice and mouse cells is recognized by several host proteins found in macrophages that are members of the ALR family and that these proteins both suppress and activate the pathways leading to the generation of cytokines and IFNs. We also show that there is great genetic diversity between different inbred mouse strains in the ALR genes, which might contribute to differential susceptibility to autoimmunity. Understanding how immune cells become activated is important to the control of disease.
Keywords: ALR; Aim2; Trex1; endogenous retrovirus; retrotransposon; self DNA.
Copyright © 2017 Nakaya et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
