A Cell Fusion-Based Screening Method Identifies Glycosylphosphatidylinositol-Anchored Protein Ly6e as the Receptor for Mouse Endogenous Retroviral Envelope Syncytin-A
- PMID: 28679758
- PMCID: PMC5571244
- DOI: 10.1128/JVI.00832-17
A Cell Fusion-Based Screening Method Identifies Glycosylphosphatidylinositol-Anchored Protein Ly6e as the Receptor for Mouse Endogenous Retroviral Envelope Syncytin-A
Abstract
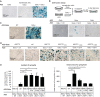
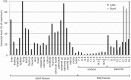
Syncytin genes are envelope genes of retroviral origin that have been exapted for a role in placentation. They are involved in the formation of a syncytial structure (the syncytiotrophoblast) at the fetomaternal interface via their fusogenic activity. The mouse placenta is unique among placental mammals since the fetomaternal interface comprises two syncytiotrophoblast layers (ST-I and ST-II) instead of one, as observed in humans and all other hemochorial placentae. Each layer specifically expresses a distinct mouse syncytin, namely, syncytin-A (SynA) for ST-I and syncytin-B (SynB) for ST-II, which have been shown to be essential to placentogenesis and embryo survival. Their cognate cellular receptors, which are necessary to mediate cell-cell fusion and syncytiotrophoblast formation, are still unknown. By devising a sensitive method that combines a cell-cell fusion assay with the screening of a mouse cDNA library, we succeeded in identifying the glycosylphosphatidylinositol (GPI)-anchored membrane protein lymphocyte antigen 6E (Ly6e) as a candidate receptor for SynA. Transfection of cells with the cloned receptor led to their fusion to cells expressing SynA, with no cross-reactive fusion activity with SynB. Knocking down Ly6e greatly reduced SynA-induced cell fusion, thus suggesting that Ly6e is the sole receptor for SynA in vivo Interaction of SynA with Ly6e was further demonstrated by a competition assay using the soluble ectodomain of Ly6e. Finally, reverse transcription-quantitative PCR (RT-qPCR) analysis of Ly6e expression on a representative panel of mouse tissues shows that it is significantly expressed in the mouse placenta together with SynA.IMPORTANCE Syncytin genes are envelope genes of endogenous retroviruses, co-opted for a physiological function in placentation. Syncytins are fusogenic proteins that mediate cell-cell fusion by interacting with receptors present on the partner cells. Here, by devising a sensitive in vitro fusion assay that enables the high-throughput screening of normalized cDNA libraries, we identified the long-sought receptor for syncytin-A (SynA), a mouse syncytin responsible for syncytiotrophoblast formation at the maternofetal interface of the mouse placenta. This protein, Ly6e (lymphocyte antigen 6E), is a GPI-anchored membrane protein, and small interfering RNA (siRNA) experiments targeting its deletion as well as a decoy assay using a recombinant soluble receptor show that Ly6e is the necessary and sufficient partner of SynA. Its profile of expression is consistent with a role in both ancestral endogenization of a SynA founder retrovirus and present-day placenta formation. This study provides a powerful general method to identify genes involved in cell-cell fusion processes.
Keywords: endogenous retrovirus; envelope protein; mouse; placenta; receptor; syncytin.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, et al. . 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi:10.1038/35057062. - DOI - PubMed
-
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, et al. . 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi:10.1038/nature01262. - DOI - PubMed
-
- Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 74:3321–3329. doi:10.1128/JVI.74.7.3321-3329.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

