Transcriptomic Profiling Reveals Differentially Expressed Genes Associated with Pine Wood Nematode Resistance in Masson Pine (Pinus massoniana Lamb.)
- PMID: 28680045
- PMCID: PMC5498564
- DOI: 10.1038/s41598-017-04944-7
Transcriptomic Profiling Reveals Differentially Expressed Genes Associated with Pine Wood Nematode Resistance in Masson Pine (Pinus massoniana Lamb.)
Abstract
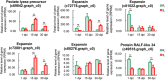
Pine wilt disease caused by pine wood nematode (Bursaphelenchus xylophilus, PWN) is a severe forest disease of the genus Pinus. Masson pine as an important timber and oleoresin resource in South China, is the major species infected by pine wilt disease. However, the underlying mechanism of pine resistance is still unclear. Here, we performed a transcriptomics analysis to identify differentially expressed genes associated with resistance to PWN infection. By comparing the expression profiles of resistant and susceptible trees inoculated with PWN at 1, 15, or 30 days post-inoculation (dpi), 260, 371 and 152 differentially expressed genes (DEGs) in resistant trees and 756, 2179 and 398 DEGs in susceptible trees were obtained. Gene Ontology enrichment analysis of DEGs revealed that the most significant biological processes were "syncytium formation" in the resistant phenotype and "response to stress" and "terpenoid biosynthesis" in the susceptible phenotype at 1 and 15 dpi, respectively. Furthermore, some key DEGs with potential regulatory roles to PWN infection, including expansins, pinene synthases and reactive oxidation species (ROS)-related genes were evaluated in detail. Finally, we propose that the biosynthesis of oleoresin and capability of ROS scavenging are pivotal to the high resistance of PWN.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Yano M. Investigation on the cause of pine mortality in Nagasaki. Prefecture sanrinkoho. 1913;4:1–14.
-
- Cheng H, Lin M, Li W, Fang Z. The occurrence of a pine wilting disease caused by a nematode found in Nanjing. For Pest Dis. 1983;4:1–5.
-
- Dwinell LD. First report of pine wood nematodes (Bursaphelenchus xylophilus) in Mexico. Plant Dis. 1993;77:846. doi: 10.1094/PD-77-0846A. - DOI
-
- Mota MM, Braasch H, Bravo MA. First record of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology. 1999;1:727–734. doi: 10.1163/156854199508757. - DOI
-
- Kobayashi F, Yamane A, Ikeda T. The Japanese pine sawyer beetle as the vector of pine wilt disease. Annu Rev Entomol. 1984;29:115–135. doi: 10.1146/annurev.en.29.010184.000555. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

