Linking functions: an additional role for an intrinsically disordered linker domain in the transcriptional coactivator CBP
- PMID: 28680062
- PMCID: PMC5498717
- DOI: 10.1038/s41598-017-04611-x
Linking functions: an additional role for an intrinsically disordered linker domain in the transcriptional coactivator CBP
Abstract
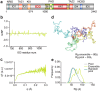
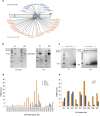
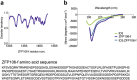
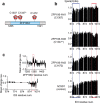
The multi-domain transcriptional coactivators CBP/p300 integrate a multitude of signaling inputs, interacting with more than 400 proteins via one or more of their globular domains. While CBP/p300 function is typically considered in terms of these structured domains, about half of the protein consists of intrinsically disordered regions (IDRs) of varying length. However, these IDRs have only been thought of as linkers that allow flexible spatial arrangement of the structured domains, but recent studies have shown that similar IDRs mediate specific and critical interactions in other proteins. To examine the roles of IDRs in CBP, we performed yeast-two-hybrid screenings of placenta and lung cancer cDNA libraries, which demonstrated that the long IDR linking the KIX domain and bromodomain of CBP (termed ID3) can potentially bind to several proteins. The RNA-binding Zinc-finger protein 106 (ZFP106) detected in both libraries was identified as a novel substrate for CBP-mediated acetylation. Nuclear magnetic resonance (NMR) spectroscopy combined with cross-linking experiments and competition-binding assays showed that the fully disordered isolated ID3 transiently interacts with an IDR of ZFP106 in a fashion that disorder of both regions is maintained. These findings demonstrate that beside the linking function, ID3 can also interact with acetylation substrates of CBP.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

